-
Notifications
You must be signed in to change notification settings - Fork 2
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Feature/results in memory #193
Conversation
* update simularium helper to use radius in molecules * update max and min radii * add size options to ingredient initialization * refactor distribution value extraction
…cellpack into feature/results-in-memory
…ture/results-in-memory
cellpack/autopack/plotly_result.py
Outdated
| @@ -59,7 +59,7 @@ def add_square(self, side_length, pos, rotMat, color, opacity=1): | |||
| ) | |||
|
|
|||
| def add_ingredient_positions(self, env): | |||
| for pos, rot, ingr, ptInd in env.molecules: | |||
| for pos, rot, ingr, ptInd, radius in env.molecules: | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
- test and fix 2D plotly results
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
- 3d sphere tree looks questionable
* formatting using black
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This was breaking image export so I made some changes to fix that. Looks good otherwise and a super useful update going forward!
| @@ -1339,11 +1242,12 @@ def get_new_pos(self, ingr, pos, rot, positions_to_adjust): | |||
| return self.transformPoints(pos, rot, positions_to_adjust) | |||
|
|
|||
| def check_against_one_packed_ingr(self, index, level, search_tree): | |||
| overlapped_ingr = self.env.rIngr[index] | |||
| packed_ingredient = self.env.packed_objects.get()[index] | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Can maybe get ingredient here directly?
| @@ -96,6 +98,28 @@ def __init__( | |||
| self.encapsulating_radius = radius | |||
| self.min_radius = radius | |||
|
|
|||
| @staticmethod | |||
| def create_voxelization( | |||
| self, image_data, bounding_box, voxel_size, image_size, position, rotation, radius, hollow=None, mesh_store=None, | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Need to test if this still works
| self, image_data, bounding_box, voxel_size, image_size, position, rotation, radius, hollow=None, mesh_store=None, | |
| image_data, bounding_box, voxel_size, image_size, position, rotation, radius, hollow=None, mesh_store=None, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I reverted this, we did this because we weren't originally storing the ingredient
| @@ -1302,7 +1302,7 @@ def displayCytoplasmIngredients(self): | |||
| else: | |||
| return | |||
| if self.doSpheres: | |||
| for pos, rot, ingr, ptInd in self.histo.molecules: | |||
| for pos, rot, ingr, ptInd, _ in self.histo.molecules: | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Are we using Graphics.py for anything? These functions are still using histo.molecules -- should get rid of them at some point.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
we're not, I can delete it in a separate simple PR
| nbFreePoints = self.onePrevIngredient( | ||
| i, mingrs, distance, nbFreePoints, organelle.molecules | ||
| ) | ||
| # TODO: refactor this to work with new placed_objects data structure |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Do we ever have object already packed while building a grid? This could also be removed if unused.
Codecov ReportAll modified lines are covered by tests ✅
❗ Your organization needs to install the Codecov GitHub app to enable full functionality. Additional details and impacted files@@ Coverage Diff @@
## main #193 +/- ##
=======================================
Coverage 98.52% 98.52%
=======================================
Files 16 16
Lines 476 476
=======================================
Hits 469 469
Misses 7 7
☔ View full report in Codecov by Sentry. |
Co-authored-by: Saurabh Mogre <[email protected]>
Co-authored-by: Saurabh Mogre <[email protected]>
Co-authored-by: Saurabh Mogre <[email protected]>
Co-authored-by: Saurabh Mogre <[email protected]>
Co-authored-by: Saurabh Mogre <[email protected]>
Co-authored-by: Saurabh Mogre <[email protected]>
…ope/cellpack into feature/results-in-memory
* use object instance instead of class to create voxelization * formatting
|
I also made a couple of minor changes to get partner packing to work with variable size since I needed it to create some synthetic data. |
…ope/cellpack into feature/results-in-memory
Co-authored-by: Saurabh Mogre <[email protected]>
…ope/cellpack into feature/results-in-memory
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
New updates look good!
- Image export is working with compartments
- 2D plots are working
- Sphere tree packing is working
|
Closes #178 |
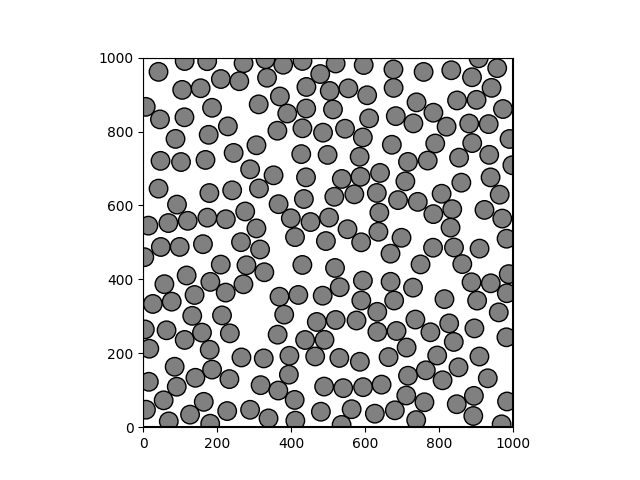

Problem
In working on the variable sized ingredient setting we realized how many different places and ways the packed data was being stored. See this wiki
Solution
We created a new class PackedObjects. At first I didn't store the ingredient in the object, but it ended up making more sense to do it (I was calling "get_ingredient_by_name" a lot).
Other changes:
compNumtocompartment_idwith @mogres
Type of change
Please delete options that are not relevant.
Steps to Verify:
pack -r '/Users/meganr/dev/mesoscope/cellpack/cellpack/tests/recipes/v2/test_variable_size.json'Screenshots (optional):