-
Notifications
You must be signed in to change notification settings - Fork 104
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Add tutorial for subcell IDP limiting (#1882)
* First version of tutorial for subcell IDP limiting * Add paragraph about time integration method * Redo code as julia comments * Hopefully fix reference * Add section about Visualizaton * Fix typos * Small changes * Add explanation about limiter, vol integral and solver * Add bounds checking section * fix typos * Add description in introduction * Clear out before hohq mesh tutorial * Adapt tutorial * Implement suggestions. * Add section about newton method and correction factor * Implement suggestion; Fix typos * Implement suggestions
- Loading branch information
Showing
4 changed files
with
301 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,287 @@ | ||
| #src # Subcell limiting with the IDP Limiter | ||
|
|
||
| # In the previous tutorial, the element-wise limiting with [`IndicatorHennemannGassner`](@ref) | ||
| # and [`VolumeIntegralShockCapturingHG`](@ref) was explained. This tutorial contains a short | ||
| # introduction to the idea and implementation of subcell shock capturing approaches in Trixi.jl, | ||
| # which is also based on the DGSEM scheme in flux differencing formulation. | ||
| # Trixi.jl contains the a-posteriori invariant domain-preserving (IDP) limiter which was | ||
| # introduced by [Pazner (2020)](https://doi.org/10.1016/j.cma.2021.113876) and | ||
| # [Rueda-Ramírez, Pazner, Gassner (2022)](https://doi.org/10.1016/j.compfluid.2022.105627). | ||
| # It is a flux-corrected transport-type (FCT) limiter and is implemented using [`SubcellLimiterIDP`](@ref) | ||
| # and [`VolumeIntegralSubcellLimiting`](@ref). | ||
| # Since it is an a-posteriori limiter you have to apply a correction stage after each Runge-Kutta | ||
| # stage. This is done by passing the stage callback [`SubcellLimiterIDPCorrection`](@ref) to the | ||
| # time integration method. | ||
|
|
||
| # ## Time integration method | ||
| # As mentioned before, the IDP limiting is an a-posteriori limiter. Its limiting process | ||
| # guarantees the target bounds for an explicit (forward) Euler time step. To still achieve a | ||
| # high-order approximation, the implementation uses strong-stability preserving (SSP) Runge-Kutta | ||
| # methods, which can be written as convex combinations of forward Euler steps. | ||
| # As such, they preserve the convexity of convex functions and functionals, such as the TVD | ||
| # semi-norm and the maximum principle in 1D, for instance. | ||
| #- | ||
| # Since IDP/FCT limiting procedure operates on independent forward Euler steps, its | ||
| # a-posteriori correction stage is implemented as a stage callback that is triggered after each | ||
| # forward Euler step in an SSP Runge-Kutta method. Unfortunately, the `solve(...)` routines in | ||
| # [OrdinaryDiffEq.jl](https://github.com/SciML/OrdinaryDiffEq.jl), typically employed for time | ||
| # integration in Trixi.jl, do not support this type of stage callback. | ||
| #- | ||
| # Therefore, subcell limiting with the IDP limiter requires the use of a Trixi-intern | ||
| # time integration SSPRK method called with | ||
| # ````julia | ||
| # Trixi.solve(ode, method(stage_callbacks = stage_callbacks); ...) | ||
| # ```` | ||
| #- | ||
| # Right now, only the canonical three-stage, third-order SSPRK method (Shu-Osher) | ||
| # [`Trixi.SimpleSSPRK33`](@ref) is implemented. | ||
|
|
||
| # # [IDP Limiting](@id IDPLimiter) | ||
| # The implementation of the invariant domain preserving (IDP) limiting approach ([`SubcellLimiterIDP`](@ref)) | ||
| # is based on [Pazner (2020)](https://doi.org/10.1016/j.cma.2021.113876) and | ||
| # [Rueda-Ramírez, Pazner, Gassner (2022)](https://doi.org/10.101/j.compfluid.2022.105627). | ||
| # It supports several types of limiting which are enabled by passing parameters individually. | ||
|
|
||
| # ### [Global bounds](@id global_bounds) | ||
| # If enabled, the global bounds enforce physical admissibility conditions, such as non-negativity | ||
| # of variables. This can be done for conservative variables, where the limiter is of a one-sided | ||
| # Zalesak-type ([Zalesak, 1979](https://doi.org/10.1016/0021-9991(79)90051-2)), and general | ||
| # non-linear variables, where a Newton-bisection algorithm is used to enforce the bounds. | ||
|
|
||
| # The Newton-bisection algorithm is an iterative method and requires some parameters. | ||
| # It uses a fixed maximum number of iteration steps (`max_iterations_newton = 10`) and | ||
| # relative/absolute tolerances (`newton_tolerances = (1.0e-12, 1.0e-14)`). The given values are | ||
| # sufficient in most cases and therefore used as default. Additionally, there is the parameter | ||
| # `gamma_constant_newton`, which can be used to scale the antidiffusive flux for the computation | ||
| # of the blending coefficients of nonlinear variables. The default value is `2 * ndims(equations)`, | ||
| # as it was shown by [Pazner (2020)](https://doi.org/10.1016/j.cma.2021.113876) [Section 4.2.2.] | ||
| # that this value guarantees the fulfillment of bounds for a forward-Euler increment. | ||
|
|
||
| # Very small non-negative values can be an issue as well. That's why we use an additional | ||
| # correction factor in the calculation of the global bounds, | ||
| # ```math | ||
| # u^{new} \geq \beta * u^{FV}. | ||
| # ``` | ||
| # By default, $\beta$ (named `positivity_correction_factor`) is set to `0.1` which works properly | ||
| # in most of the tested setups. | ||
|
|
||
| # #### Conservative variables | ||
| # The procedure to enforce global bounds for a conservative variables is as follows: | ||
| # If you want to guarantee non-negativity for the density of the compressible Euler equations, | ||
| # you pass the specific quantity name of the conservative variable. | ||
| using Trixi | ||
| equations = CompressibleEulerEquations2D(1.4) | ||
|
|
||
| # The quantity name of the density is `rho` which is how we enable its limiting. | ||
| positivity_variables_cons = ["rho"] | ||
|
|
||
| # The quantity names are passed as a vector to allow several quantities. | ||
| # This is used, for instance, if you want to limit the density of two different components using | ||
| # the multicomponent compressible Euler equations. | ||
| equations = CompressibleEulerMulticomponentEquations2D(gammas = (1.4, 1.648), | ||
| gas_constants = (0.287, 1.578)) | ||
|
|
||
| # Then, we just pass both quantity names. | ||
| positivity_variables_cons = ["rho1", "rho2"] | ||
|
|
||
| # Alternatively, it is possible to all limit all density variables with a general command using | ||
| positivity_variables_cons = ["rho" * string(i) for i in eachcomponent(equations)] | ||
|
|
||
| # #### Non-linear variables | ||
| # To allow limitation for all possible non-linear variables, including variables defined | ||
| # on-the-fly, you can directly pass the function that computes the quantity for which you want | ||
| # to enforce positivity. For instance, if you want to enforce non-negativity for the pressure, | ||
| # do as follows. | ||
| positivity_variables_nonlinear = [pressure] | ||
|
|
||
| # ### Local bounds | ||
| # Second, Trixi.jl supports the limiting with local bounds for conservative variables using a | ||
| # two-sided Zalesak-type limiter ([Zalesak, 1979](https://doi.org/10.1016/0021-9991(79)90051-2)). | ||
| # They allow to avoid spurious oscillations within the global bounds and to improve the | ||
| # shock-capturing capabilities of the method. The corresponding numerical admissibility conditions | ||
| # are frequently formulated as local maximum or minimum principles. The local bounds are computed | ||
| # using the maximum and minimum values of all local neighboring nodes. Within this calculation we | ||
| # use the low-order FV solution values for each node. | ||
|
|
||
| # As for the limiting with global bounds you are passing the quantity names of the conservative | ||
| # variables you want to limit. So, to limit the density with lower and upper local bounds pass | ||
| # the following. | ||
| local_minmax_variables_cons = ["rho"] | ||
|
|
||
| # ## Exemplary simulation | ||
| # How to set up a simulation using the IDP limiting becomes clearer when looking at an exemplary | ||
| # setup. This will be a simplified version of `tree_2d_dgsem/elixir_euler_blast_wave_sc_subcell.jl`. | ||
| # Since the setup is mostly very similar to a pure DGSEM setup as in | ||
| # `tree_2d_dgsem/elixir_euler_blast_wave.jl`, the equivalent parts are used without any explanation | ||
| # here. | ||
| using OrdinaryDiffEq | ||
| using Trixi | ||
|
|
||
| equations = CompressibleEulerEquations2D(1.4) | ||
|
|
||
| function initial_condition_blast_wave(x, t, equations::CompressibleEulerEquations2D) | ||
| ## Modified From Hennemann & Gassner JCP paper 2020 (Sec. 6.3) -> "medium blast wave" | ||
| ## Set up polar coordinates | ||
| inicenter = SVector(0.0, 0.0) | ||
| x_norm = x[1] - inicenter[1] | ||
| y_norm = x[2] - inicenter[2] | ||
| r = sqrt(x_norm^2 + y_norm^2) | ||
| phi = atan(y_norm, x_norm) | ||
| sin_phi, cos_phi = sincos(phi) | ||
|
|
||
| ## Calculate primitive variables | ||
| rho = r > 0.5 ? 1.0 : 1.1691 | ||
| v1 = r > 0.5 ? 0.0 : 0.1882 * cos_phi | ||
| v2 = r > 0.5 ? 0.0 : 0.1882 * sin_phi | ||
| p = r > 0.5 ? 1.0E-3 : 1.245 | ||
|
|
||
| return prim2cons(SVector(rho, v1, v2, p), equations) | ||
| end | ||
| initial_condition = initial_condition_blast_wave; | ||
|
|
||
| # Since the surface integral is equal for both the DG and the subcell FV method, the limiting is | ||
| # applied only in the volume integral. | ||
| #- | ||
| # Note, that the DG method is based on the flux differencing formulation. Hence, you have to use a | ||
| # two-point flux, such as [`flux_ranocha`](@ref), [`flux_shima_etal`](@ref), [`flux_chandrashekar`](@ref) | ||
| # or [`flux_kennedy_gruber`](@ref), for the DG volume flux. | ||
| surface_flux = flux_lax_friedrichs | ||
| volume_flux = flux_ranocha | ||
|
|
||
| # The limiter is implemented within [`SubcellLimiterIDP`](@ref). It always requires the | ||
| # parameters `equations` and `basis`. With additional parameters (described [above](@ref IDPLimiter) | ||
| # or listed in the docstring) you can specify and enable additional limiting options. | ||
| # Here, the simulation should contain local limiting for the density using lower and upper bounds. | ||
| basis = LobattoLegendreBasis(3) | ||
| limiter_idp = SubcellLimiterIDP(equations, basis; | ||
| local_minmax_variables_cons = ["rho"]) | ||
|
|
||
| # The initialized limiter is passed to `VolumeIntegralSubcellLimiting` in addition to the volume | ||
| # fluxes of the low-order and high-order scheme. | ||
| volume_integral = VolumeIntegralSubcellLimiting(limiter_idp; | ||
| volume_flux_dg = volume_flux, | ||
| volume_flux_fv = surface_flux) | ||
|
|
||
| # Then, the volume integral is passed to `solver` as it is done for the standard flux-differencing | ||
| # DG scheme or the element-wise limiting. | ||
| solver = DGSEM(basis, surface_flux, volume_integral) | ||
| #- | ||
| coordinates_min = (-2.0, -2.0) | ||
| coordinates_max = (2.0, 2.0) | ||
| mesh = TreeMesh(coordinates_min, coordinates_max, | ||
| initial_refinement_level = 5, | ||
| n_cells_max = 10_000) | ||
|
|
||
| semi = SemidiscretizationHyperbolic(mesh, equations, initial_condition, solver) | ||
|
|
||
| tspan = (0.0, 2.0) | ||
| ode = semidiscretize(semi, tspan) | ||
|
|
||
| summary_callback = SummaryCallback() | ||
|
|
||
| analysis_interval = 1000 | ||
| analysis_callback = AnalysisCallback(semi, interval = analysis_interval) | ||
|
|
||
| alive_callback = AliveCallback(analysis_interval = analysis_interval) | ||
|
|
||
| save_solution = SaveSolutionCallback(interval = 1000, | ||
| save_initial_solution = true, | ||
| save_final_solution = true, | ||
| solution_variables = cons2prim) | ||
|
|
||
| stepsize_callback = StepsizeCallback(cfl = 0.3) | ||
|
|
||
| callbacks = CallbackSet(summary_callback, | ||
| analysis_callback, alive_callback, | ||
| save_solution, | ||
| stepsize_callback); | ||
|
|
||
| # As explained above, the IDP limiter works a-posteriori and requires the additional use of a | ||
| # correction stage implemented with the stage callback [`SubcellLimiterIDPCorrection`](@ref). | ||
| # This callback is passed within a tuple to the time integration method. | ||
| stage_callbacks = (SubcellLimiterIDPCorrection(),) | ||
|
|
||
| # Moreover, as mentioned before as well, simulations with subcell limiting require a Trixi-intern | ||
| # SSPRK time integration methods with passed stage callbacks and a Trixi-intern `Trixi.solve(...)` | ||
| # routine. | ||
| sol = Trixi.solve(ode, Trixi.SimpleSSPRK33(stage_callbacks = stage_callbacks); | ||
| dt = 1.0, # solve needs some value here but it will be overwritten by the stepsize_callback | ||
| callback = callbacks); | ||
| summary_callback() # print the timer summary | ||
|
|
||
|
|
||
| # ## Visualization | ||
| # As for a standard simulation in Trixi.jl, it is possible to visualize the solution using the | ||
| # `plot` routine from Plots.jl. | ||
| using Plots | ||
| plot(sol) | ||
|
|
||
| # To get an additional look at the amount of limiting that is used, you can use the visualization | ||
| # approach using the [`SaveSolutionCallback`](@ref), [`Trixi2Vtk`](https://github.com/trixi-framework/Trixi2Vtk.jl) | ||
| # and [ParaView](https://www.paraview.org/download/). More details about this procedure | ||
| # can be found in the [visualization documentation](@ref visualization). | ||
| # Unfortunately, the support for subcell limiting data is not yet merged into the main branch | ||
| # of Trixi2Vtk but lies in the branch [`bennibolm/node-variables`](https://github.com/bennibolm/Trixi2Vtk.jl/tree/node-variables). | ||
| #- | ||
| # With that implementation and the standard procedure used for Trixi2Vtk you get the following | ||
| # dropdown menu in ParaView. | ||
| #- | ||
| #  | ||
|
|
||
| # The resulting visualization of the density and the limiting parameter then looks like this. | ||
| # 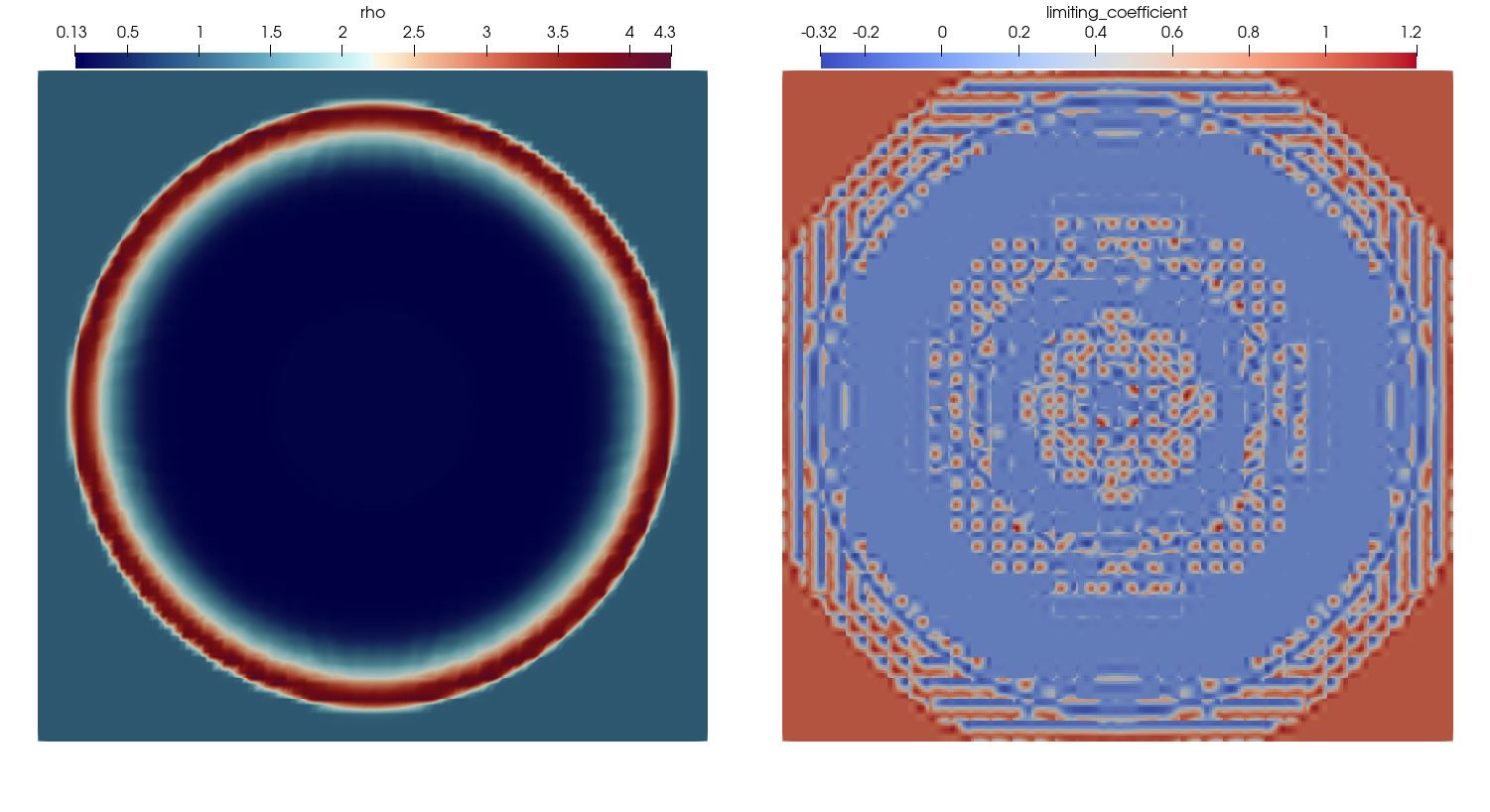 | ||
|
|
||
| # You can see that the limiting coefficient does not lie in the interval [0,1] because Trixi2Vtk | ||
| # interpolates all quantities to regular nodes by default. | ||
| # You can disable this functionality with `reinterpolate=false` within the call of `trixi2vtk(...)` | ||
| # and get the following visualization. | ||
| # 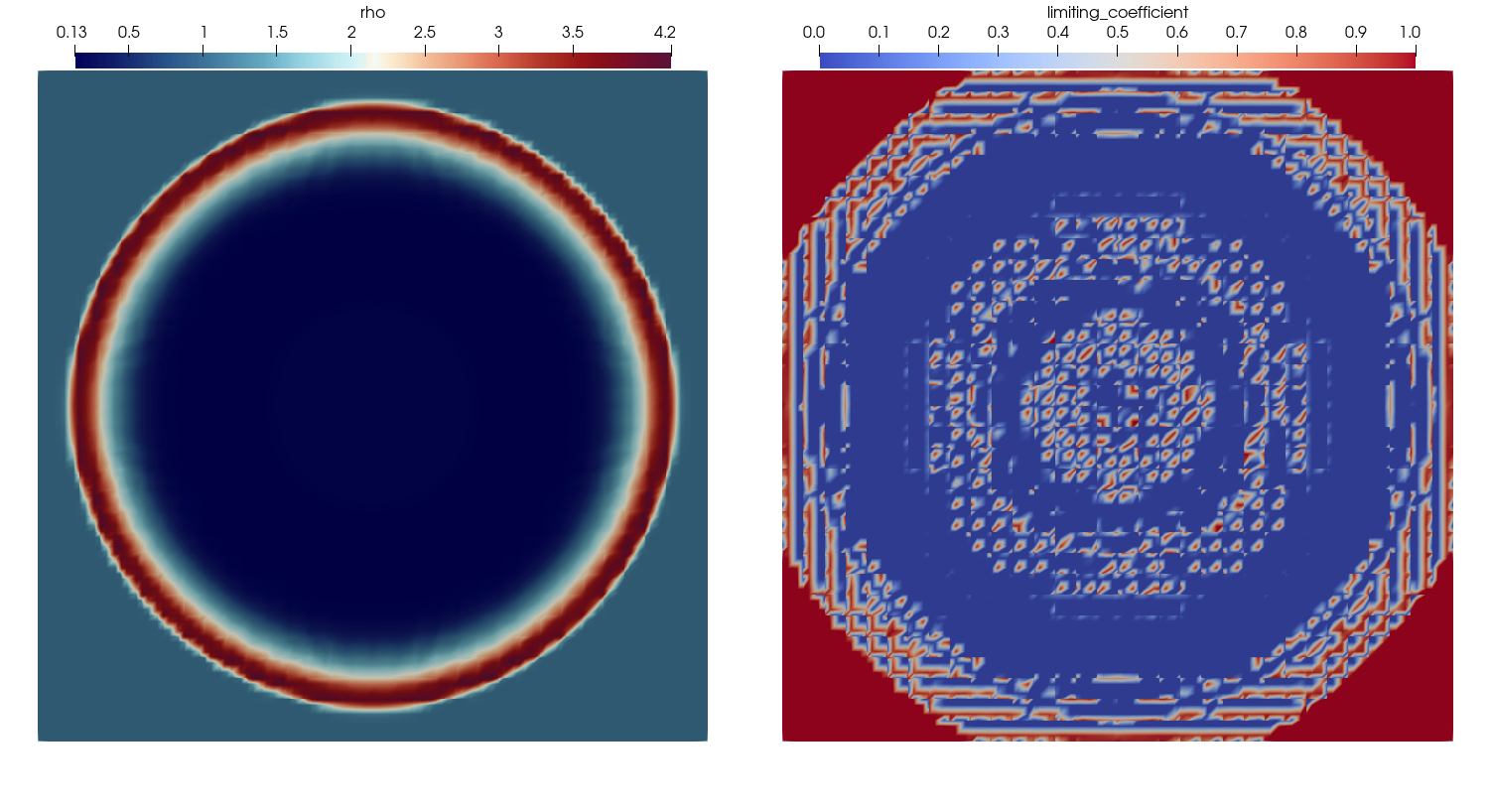 | ||
|
|
||
|
|
||
| # ## Bounds checking | ||
| # Subcell limiting is based on the fulfillment of target bounds - either global or local. | ||
| # Although the implementation works and has been thoroughly tested, there are some cases where | ||
| # these bounds are not met. | ||
| # For instance, the deviations could be in machine precision, which is not problematic. | ||
| # Larger deviations can be cause by too large time-step sizes (which can be easily fixed by | ||
| # reducing the CFL number), specific boundary conditions or source terms. Insufficient parameters | ||
| # for the Newton-bisection algorithm can also be a reason when limiting non-linear variables. | ||
| # There are described [above](@ref global_bounds). | ||
| #- | ||
| # In many cases, it is reasonable to monitor the bounds deviations. | ||
| # Because of that, Trixi.jl supports a bounds checking routine implemented using the stage | ||
| # callback [`BoundsCheckCallback`](@ref). It checks all target bounds for fulfillment | ||
| # in every RK stage. If added to the tuple of stage callbacks like | ||
| # ````julia | ||
| # stage_callbacks = (SubcellLimiterIDPCorrection(), BoundsCheckCallback()) | ||
| # ```` | ||
| # and passed to the time integration method, a summary is added to the final console output. | ||
| # For the given example, this summary shows that all bounds are met at all times. | ||
| # ```` | ||
| # ──────────────────────────────────────────────────────────────────────────────────────────────────── | ||
| # Maximum deviation from bounds: | ||
| # ──────────────────────────────────────────────────────────────────────────────────────────────────── | ||
| # rho: | ||
| # - lower bound: 0.0 | ||
| # - upper bound: 0.0 | ||
| # ──────────────────────────────────────────────────────────────────────────────────────────────────── | ||
| # ```` | ||
|
|
||
| # Moreover, it is also possible to monitor the bounds deviations incurred during the simulations. | ||
| # To do that use the parameter `save_errors = true`, such that the instant deviations are written | ||
| # to `deviations.txt` in `output_directory` every `interval` time steps. | ||
| # ````julia | ||
| # BoundsCheckCallback(save_errors = true, output_directory = "out", interval = 100) | ||
| # ```` | ||
| # Then, for the given example the deviations file contains all daviations for the current | ||
| # timestep and simulation time. | ||
| # ```` | ||
| # iter, simu_time, rho_min, rho_max | ||
| # 1, 0.0, 0.0, 0.0 | ||
| # 101, 0.29394033217556337, 0.0, 0.0 | ||
| # 201, 0.6012597465597065, 0.0, 0.0 | ||
| # 301, 0.9559096690030839, 0.0, 0.0 | ||
| # 401, 1.3674274981949077, 0.0, 0.0 | ||
| # 501, 1.8395301696603052, 0.0, 0.0 | ||
| # 532, 1.9974179806990118, 0.0, 0.0 | ||
| # ```` |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters