This repository has been archived by the owner on Nov 17, 2017. It is now read-only.
-
Notifications
You must be signed in to change notification settings - Fork 14
Home
Keiichiro Ono edited this page Jun 18, 2015
·
84 revisions
Build and share your own Cytoscape workflows with R, Python, and all other popular tools
- 6/14/2015: 0.9.17 released. Layout attributes and local table columns are supported.
- 6/9/2015: 0.9.16 released. Now Layout parameters are accessible through API.
- 6/5/2015: Python wrapper (py2cytoscape) released.
With cyREST, you can:
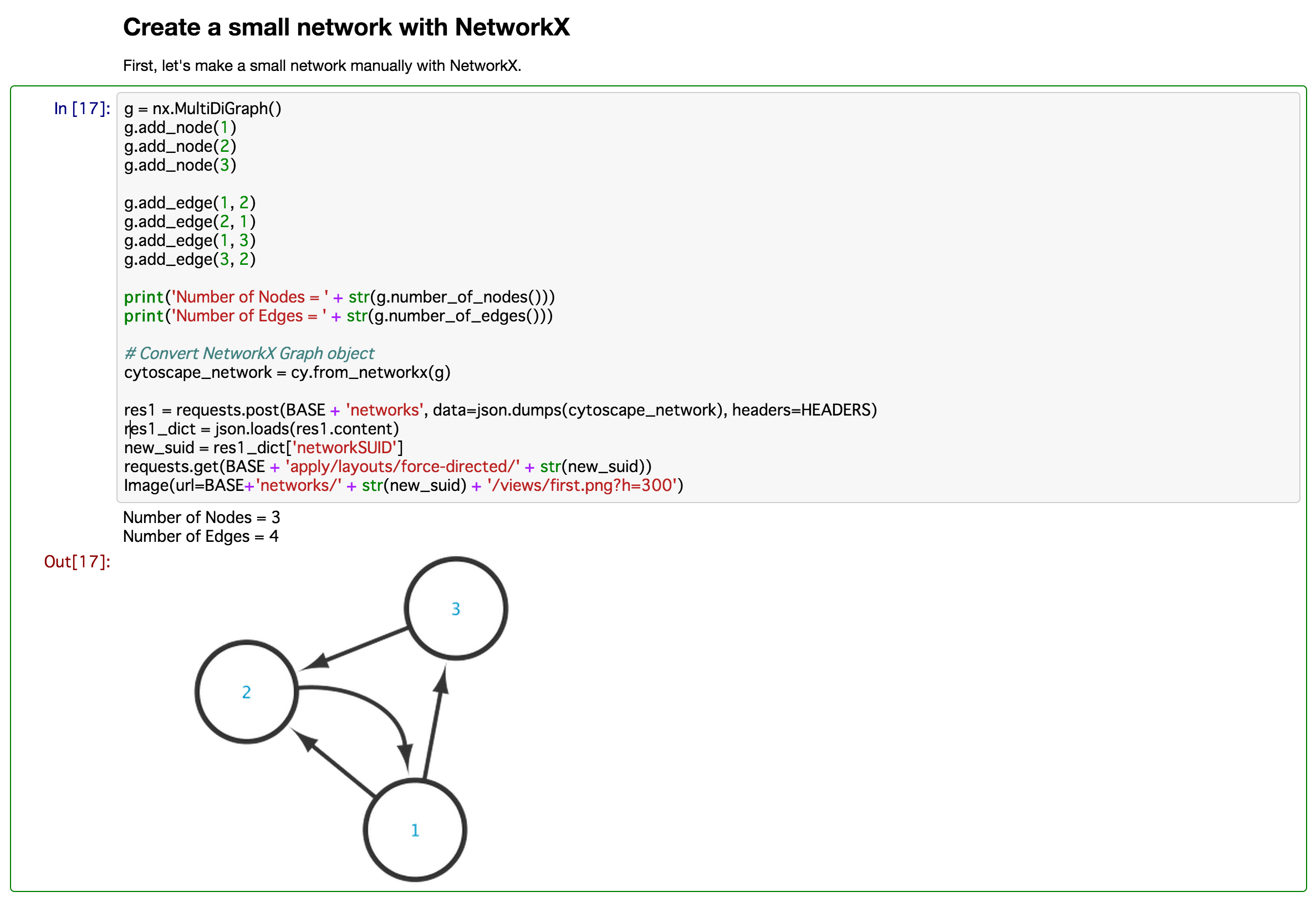
- Access Cytoscape data objects from IPython/Jupyter Notebook
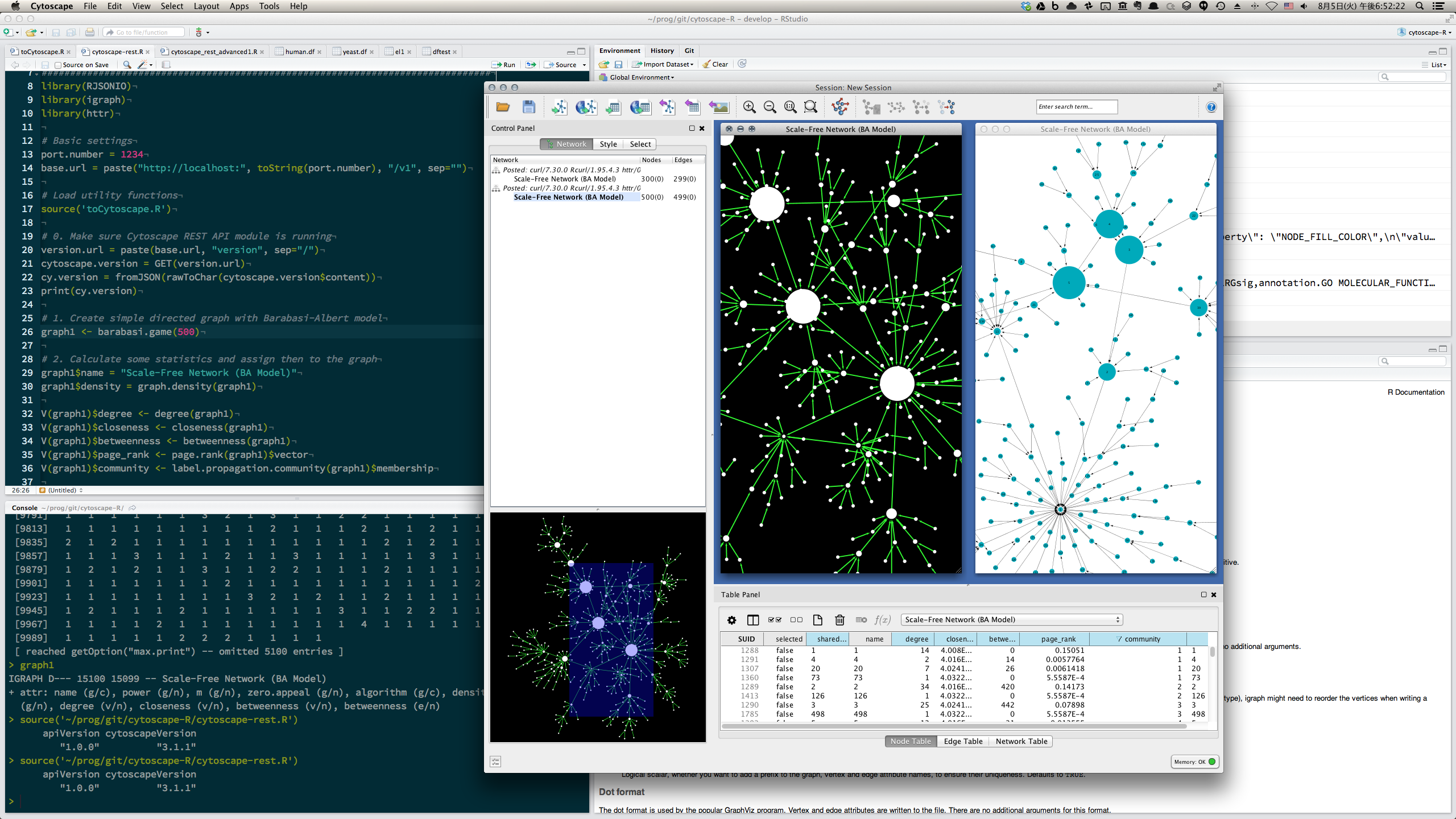
- Directly visualize network data stored as igraph object in R session
- Script your workflow in your favorite programming languages

- Python
- R
- JavaScript (Node.js)
- (More coming soon...)
(TBD)
Please send us your questions and comments to our mailing list.
- Source Code: The MIT license
- Documentation: CC BY-SA 4.0
© 2014-2015 The Cytoscape Consortium
Developed and Maintained by Keiichiro Ono (UC, San Diego Trey Ideker Lab)