diff --git a/README.md b/README.md
index b236c6d7..a8c64248 100644
--- a/README.md
+++ b/README.md
@@ -59,6 +59,8 @@ Geochemistry π was selected for featuring as an Editor’s Highlight in EOS
Eos Website: https://eos.org/editor-highlights/machine-learning-for-geochemists-who-dont-want-to-code.
+
+
## Quick Installation
Our software is well tested on **macOS** and **Windows** system with **Python 3.9**. Other systems and Python version are not guranteed.
diff --git a/docs/source/For Developer/Add New Model To Framework.md b/docs/source/For Developer/Add New Model To Framework.md
index 3b3eb0c2..d2276ea9 100644
--- a/docs/source/For Developer/Add New Model To Framework.md
+++ b/docs/source/For Developer/Add New Model To Framework.md
@@ -1,276 +1,230 @@
# Add New Model To Framework
+## Table of Contents
+- [1. Framework - Design Pattern and Hierarchical Pipeline Architecture](#1-design-pattern)
+- [2. Understand Machine Learning Algorithm](#2-understand-ml)
+- [3. Construct Model Workflow Class](#3-construct-model)
+ - [3.1 Add Basic Elements](#3-1-add-basic-element)
+ - [3.1.1 Find File](#3-1-1-find-file)
+ - [3.1.2 Define Class Attributes and Constructor](#3-1-2-define-class-attributes-and-constructors)
+ - [3.2 Add Manual Hyperparameter Tuning Functionality](#3-2-add-manual)
+ - [3.2.1 Define manual_hyper_parameters Method](#3-2-1-define-manaul-method)
+ - [3.2.2 Create _algorithm.py File](#3-2-2-create-file)
+ - [3.3 Add Automated Hyperparameter Tuning (AutoML) Functionality](#3-3-add-automl)
+ - [3.3.1 Add AutoML Code to Model Workflow Class](#3-3-1-add-automl-code)
+ - [3.4 Add Application Function to Model Workflow Class](#3-4-add-application-function)
+ - [3.4.1 Add Common Application Functions and common_components Method](#3-4-1-add-common-function)
+ - [3.4.2 Add Special Application Functions and special_components Method](#3-4-2-add-special-function)
+ - [3.4.3 Add @dispatch() to Component Method](#3-4-3-add-dispatch)
+ - [3.5 Storage Mechanism](#3-5-storage-mechanism)
+- [4. Instantiate Model Workflow Class](#4-instantiate-model-workflow-class)
+ - [4.1 Find File](#4-1-find-file)
+ - [4.2 Import Module](#4-2-import-module)
+ - [4.3 Define activate Method](#4-3-define-activate-method)
+ - [4.4 Create Model Workflow Object](#4-4-create-model-workflow-object)
+ - [4.5 Invoke Other Methods in Scikit-learn API Style](#4-5-invoke-other-methods)
+ - [4.6 Add model_name to MODE_MODELS or NON_AUTOML_MODELS](#4-6-add-model-name)
+- [5. Test Model Workflow Class](#5-test-model)
+- [6. Completed Pull Request](#6-completed-pull-request)
+- [7. Precautions](#7-precautions)
-## Table of Contents
+## 1. Framework - Design Pattern and Hierarchical Pipeline Architecture
+
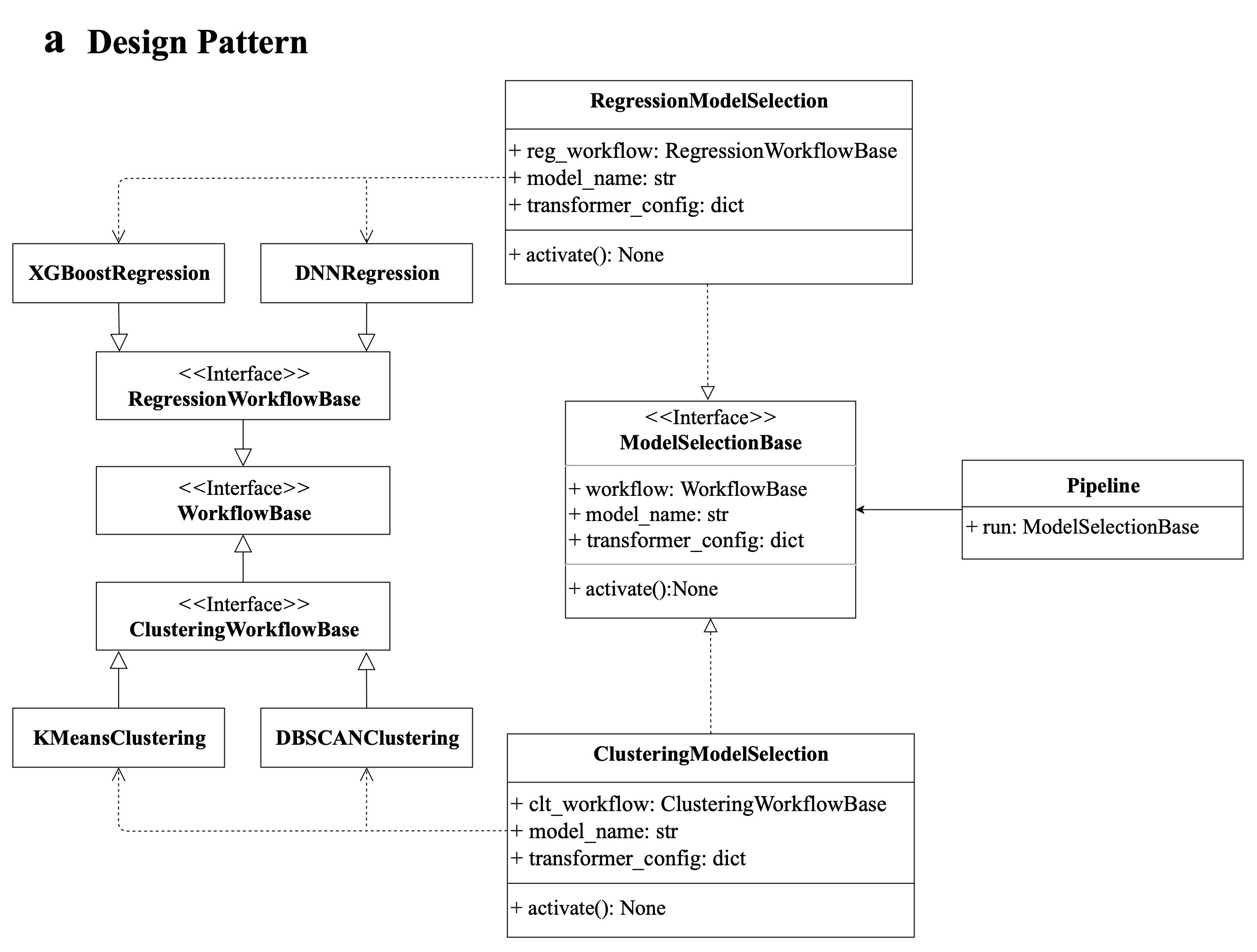
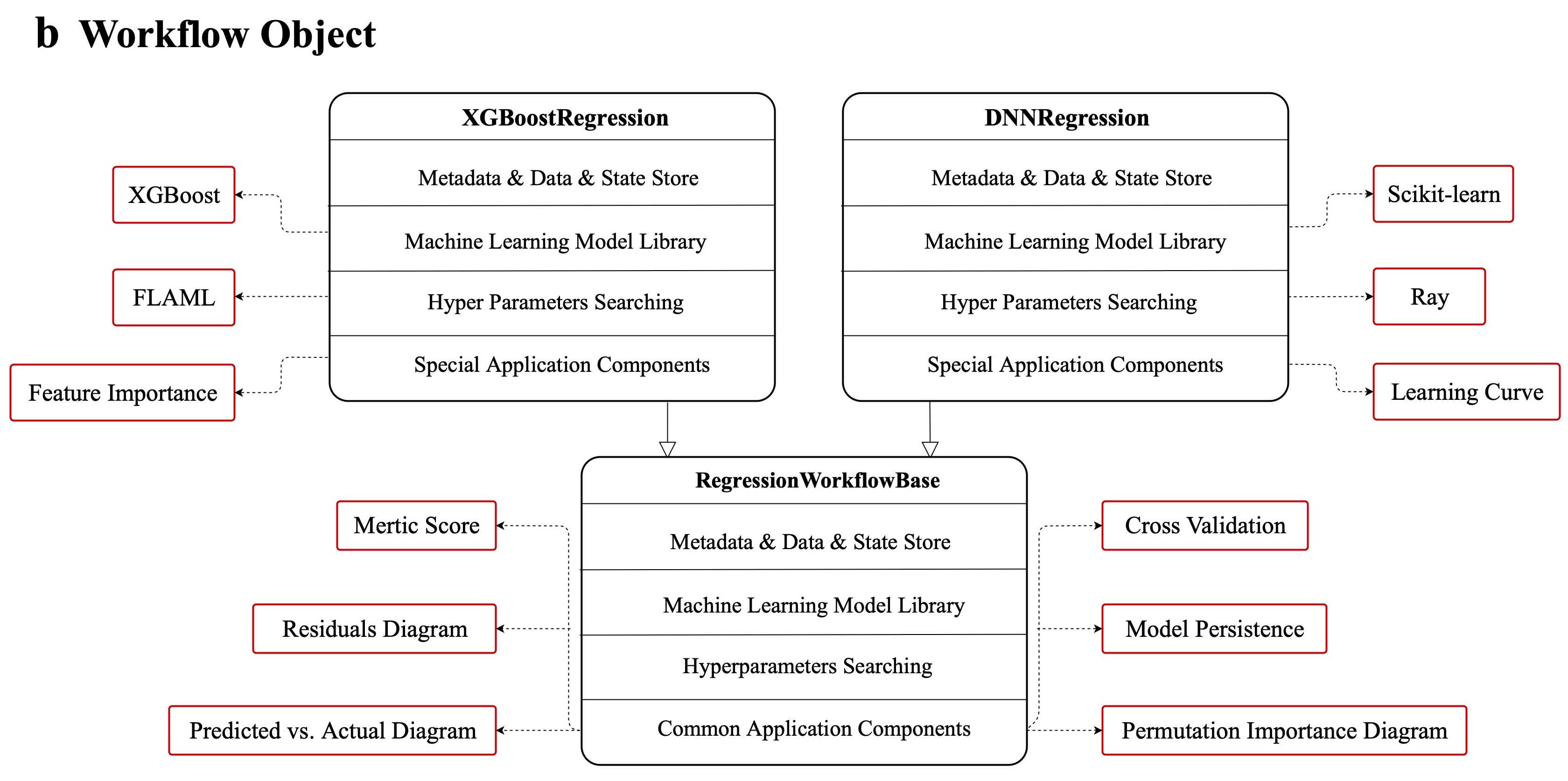
+Geochemistry π refers to the software design pattern "Abstract Factory", serving as the foundational framework upon which our advanced automated ML capabilities are built.
+
+
+
-- [1. Understand the model](#1-understand-the-model)
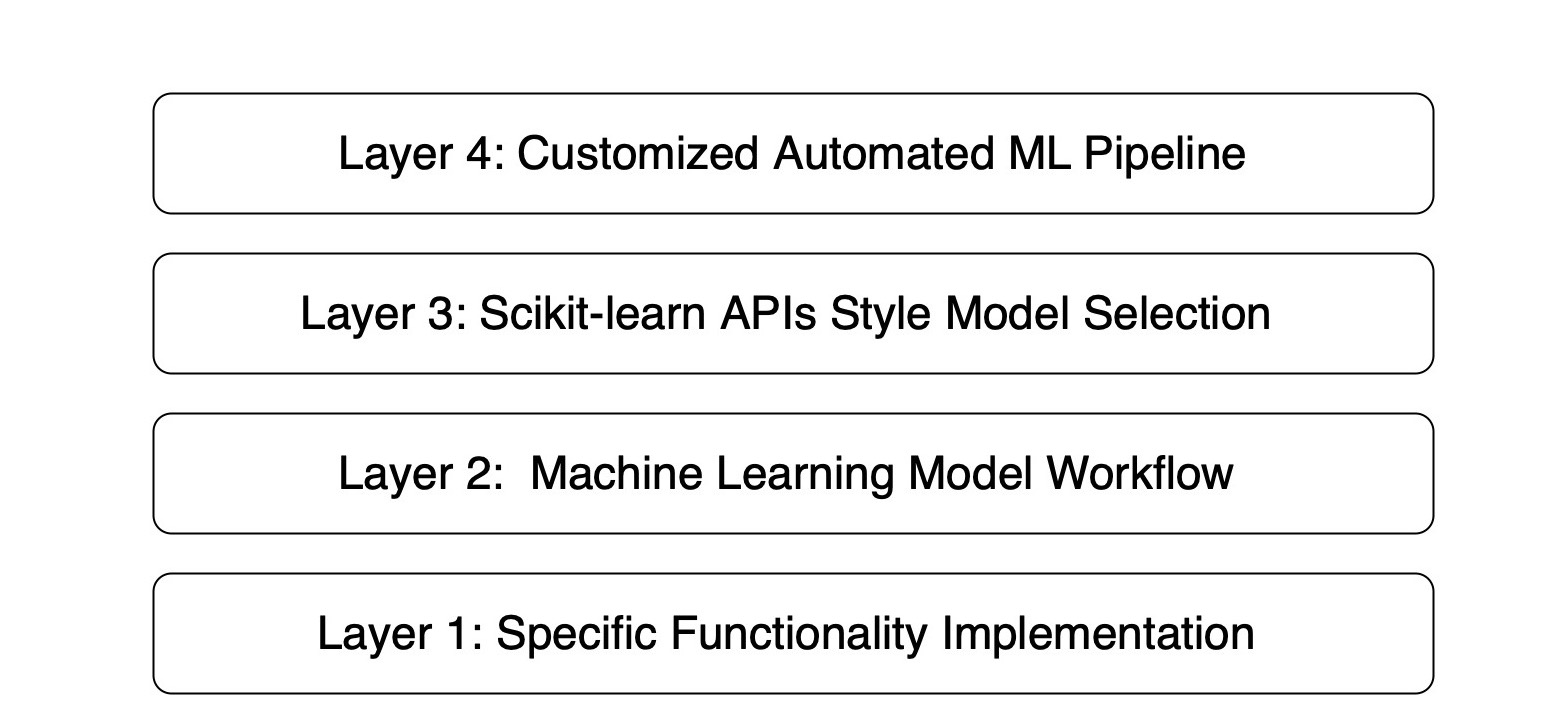
+The framework is a four-layer hierarchical pipeline architecture that promotes the creation of workflow obiects through a set of model selection interfaces. The critical layers of this architecture are, as follows:
-- [2. Add Model](#2-add-model)
- - [2.1 Add The Model Class](#21-add-the-model-class)
- - [2.1.1 Find Add File](#211-find-add-file)
- - [2.1.2 Define class properties and constructors, etc.](#212-define-class-properties-and-constructors-etc)
- - [2.1.3 Define manual\_hyper\_parameters](#213-define-manual_hyper_parameters)
- - [2.1.4 Define special\_components](#214-define-special_components)
+1. Layer 1: the realization of ML model-associated functionalities with specific dependencies or libraries.
+2. Layer 2: the abstract components of the ML model workflow class include regression, classification, clustering, and decomposition.
+3. Layer 3: the scikit-learn API-style model selection interface implements the creation of ML model workflow objects.
+4. Layer 4: the customized automated ML pipeline operated at the command line or through a web interface with a complete data-mining process.
- - [2.2 Add AutoML](#22-add-automl)
- - [2.2.1 Add AutoML code to class](#221-add-automl-code-to-class)
+
+  +
+
- - [2.3 Get the hyperparameter value through interactive methods](#23-get-the-hyperparameter-value-through-interactive-methods)
- - [2.3.1 Find file](#231-find-file)
- - [2.3.2 Create the .py file and add content](#232-create-the-py-file-and-add-content)
- - [2.3.3 Import in the file that defines the model class](#233-import-in-the-file-that-defines-the-model-class)
+This pattern-driven architecture offers developers a standardized and intuitive way to create a ML model workflow class in Layer 2 by using a unified and consistent approach to object creation in Layer 3. Furthermore, it ensures the interchangeability of different model applications, allowing for seamless transitions between methodologies in Layer 1.
- - [2.4 Call Model](#24-call-model)
- - [2.4.1 Find file](#241-find-file)
- - [2.4.2 Import module](#242-import-module)
- - [2.4.3 Call model](#243-call-model)
+ - - [2.5 Add the algorithm list and set NON\_AUTOML\_MODELS](#25-add-the-algorithm-list-and-set-non_automl_models)
- - [2.5.1 Find file](#251-find-file)
+The code of each layer lies as shown above.
- - [2.6 Add Functionality](#26-add-functionality)
+**Notice**: in our framework, a **model workflow class** refers to an **algorithm workflow class** and a **mode** includes multiple model workflow classes.
- - [2.6.1 Model Research](#261-model-research)
+Now, we will take KMeans algorithm as an example to illustrate the connection between each layer. Don't get too hung up on ths part. Once you finish reading the whole article, you can come back to here again.
- - [2.6.2 Add Common_component](#262-add-common_component)
+After reading this article, you are recommended to refer to this publication also for more details on the whole scope of our framework:
- - [2.6.3 Add Special_component](#263-add-special_component)
+https://agupubs.onlinelibrary.wiley.com/doi/10.1029/2023GC011324
-- [3. Test model](#3-test-model)
-- [4. Completed Pull Request](#4-completed-pull-request)
+## 2. Understand Machine Learning Algorithm
+You need to understand the general meaning of the machine learning algorithm you are responsible for. Then you encapsultate it as an algorithm workflow in our framework and put it under the directory `geochemistrypi/data_mining/model`. Then you need to determine which **mode** this algorithm belongs to and the role of each parameter. For example, linear regression algorithm belongs to regression mode in our framework.
-- [5. Precautions](#5-precautions)
++ When learning the ML algorithm, you can refer to the relevant knowledge on the [scikit-learn official website](https://scikit-learn.org/stable/index.html).
-## 1. Understand the model
-You need to understand the general meaning of the model, determine which algorithm the model belongs to and the role of each parameter.
-+ You can choose to learn about the relevant knowledge on the [scikit-learn official website](https://scikit-learn.org/stable/index.html).
+## 3. Construct Model Workflow Class
+**Noted**: You can reference any existing model workflow classes in our framework to implement your own model workflow class.
-## 2. Add Model
-### 2.1 Add The Model Class
-#### 2.1.1 Find Add File
-First, you need to define the model class that you need to complete in the corresponding algorithm file. The corresponding algorithm file is in the `model` folder in the `data_mining` folder in the `geochemistrypi` folder.
+### 3.1 Add Basic Elements
+
+#### 3.1.1 Find File
+First, you need to construct the algorithm workflow class in the corresponding model file. The corresponding model file locates under the path `geochemistrypi/data_mining/model`.
-**eg:** If you want to add a model to the regression algorithm, you need to add it in the `regression.py` file.
+**E.g.,** If you want to add a model for the regression mode, you need to add it in the `regression.py` file.
+
+#### 3.1.2 Define Class Attributes and Constructor
+
+(1) Define the algorithm workflow class and its base class
-#### 2.1.2 Define class properties and constructors, etc.
-(1) Define the class and the required Base class
```
-class NAME (Base class):
+class ModelWorkflowClassName(BaseModelWorkflowClassName):
```
-+ NAME is the name of the algorithm, you can refer to the NAME of other models, the format needs to be consistent.
-+ Base class needs to be selected according to the actual algorithm requirements.
++ You can refer to the ModelName of other models, the format (Upper case and with the suffix 'Corresponding Mode') needs to be consistent. E.g., `XGBoostRegression`.
++ Base class needs to be inherited according to the mode the model belongs to.
```
-"""The automation workflow of using "Name" to make insightful products."""
+"""The automation workflow of using "ModelWorkflowClassName" algorithm to make insightful products."""
```
-+ Class explanation, you can refer to other classes.
++ Class docstring, you can refer to other classes. The template is shown above.
-(2) Define the name and the special_function
+(2) Define the class attributes `name`
```
-name = "name"
+name = "algorithm terminology"
```
-+ Define name, different from NMAE.
-+ This name needs to be added to the _`constants.py`_ file and the corresponding algorithm file in the `process` folder. Note that the names are consistent.
++ The class attributes `name` is different from ModelWorkflowClassName. E.g., the name `XGBoost` in `XGBoostRegression` model workflow class.
++ This name needs to be added to the corresponding constant variable in `geochemistrypi/data_mining/constants.py` file and the corresponding mode processing file under the `geochemistrypi/data_mining/process` folder. Note that those name value should be identical. It will be further explained in later section.
++ For example, the name value `XGBoost` should be included in the constant varible `REGRESSION_MODELS` in `geochemistrypi/data_mining/constants.py` file and it will be use in `geochemistrypi/data_mining/process/regress.py`.
+
+(3) Define the class attrbutes `common_functiion` or `special_function`
+
+If this model workflow class is a base class, you need to define the class attrbutes `common_functiion`. For example, the class attrbutes `common_functiion` in the base workflow class `RegressionWorkflowBase`.
+
+The values of `common_functiion` are the description of the functionalities of the models belonging to the same mode. It means the children class (all regession models) can share the same common functionalies as well.
+
+```
+common_functiion = []
+```
+
+If this model workflow class is a specific model workflow class, you need to define the class attrbutes `special_function`. For example, the class attrbutes `special_function` in the model workflow class `XGBoostRegression`.
+
+The values of `special_function` are the description of the owned functionalities of that specific model. Those special functions cannot be reused by other models.
+
```
special_function = []
```
-+ special_function is added according to the specific situation of the model, you can refer to other similar models.
-(3) Define constructor
+More detail will be explained in the later section.
+
+(4) Define the signature of the constructor
```
def __init__(
self,
- parameter:type=Default parameter value,
+ parameter: type = Default parameter value,
) -> None:
```
-+ All parameters in the corresponding model function need to be written out.
-+ Default parameter value needs to be set according to official documents.
++ The parameters in the constructor is from the algorithm library you depend on. For example, you use **Lasso** algorithm from Sckit-learn library. You can reference its introduction ([Lasso](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html)) in Scikit-learn website.
++ Default parameter value needs to be set according to scikit-learn official documents also.
+
+
+
```
- """
+"""
Parameters
----------
-parameter:type,default = Dedault
+parameter: type,default = Dedault
References
----------
Scikit-learn API: sklearn.model.name
https://scikit-learn.org/......
```
-+ Parameters is in the source of the corresponding model on the official website of sklearn
-
-**eg:** Take the [Lasso algorithm](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html) as a column.
-
-
++ Parameters docstring are in the source code of the corresponding algorithm on the official website of sklearn.
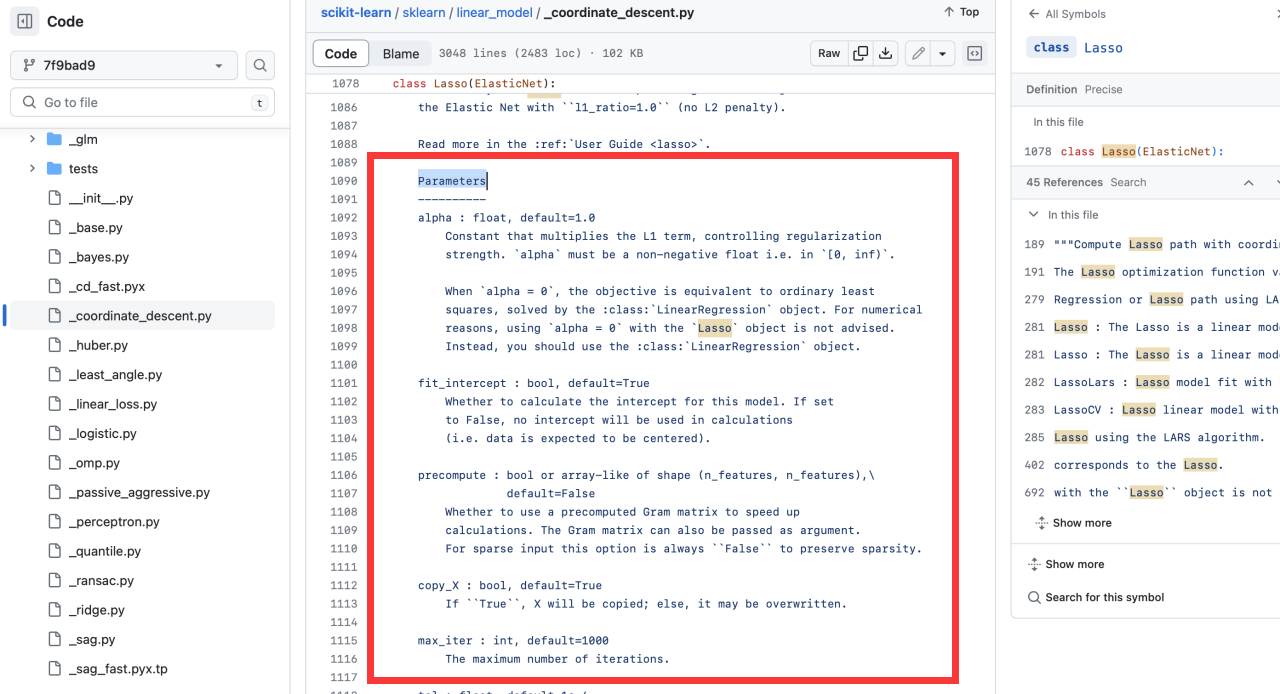
-+ References is your model's official website.
-(4) The constructor of Base class is called
+(5) The constructor of Base class is called
```
super().__init__()
```
-(5) Initializes the instance's state by assigning the parameter values passed to the constructor to the instance's properties.
+
+(6) Initializes the instance's state by assigning the parameter values passed to the constructor to the instance's attributes.
```
self.parameter=parameter
```
-(6) Create the model and assign
+
+(7) Instantiate the algorithm class you depend on and assign. For example, `Lasso` from the library `sklearn.linear_model`.
```
-self.model=modelname(
+self.model = modelname(
parameter=self.parameter
)
```
-**Note:** Don't forget to import Model from scikit-learn
+**Note:** Don't forget to import the class from scikit-learn library
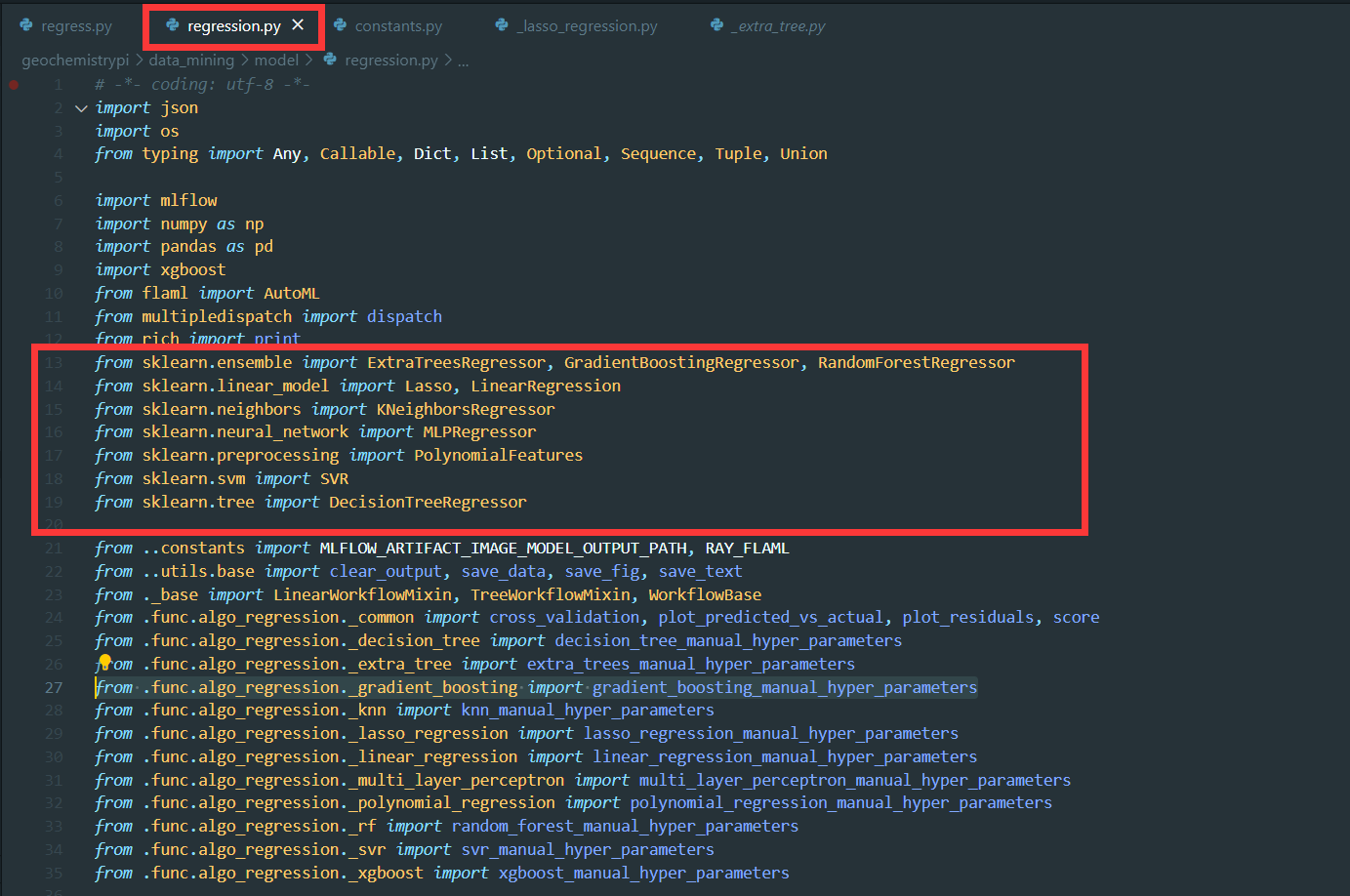
-(7) Define other class properties
+(8) Define the instance attribute `naming`
```
-self.properties=...
-```
-
-#### 2.1.3 Define manual_hyper_parameters
-manual_hyper_parameters gets the hyperparameter value by calling the manual hyperparameter function, and returns hyper_parameters.
+self.naming = Class.name
```
-hyper_parameters = name_manual_hyper_parameters()
-```
-+ This function calls the corresponding function in the `func` folder (needs to be written, see 2.2.2) to get the hyperparameter value.
-
-+ This function is called in the corresponding file of the `Process` folder (need to be written, see 2.3).
-+ Can be written with reference to similar classes
-
+This one will be use to print the name of the class and to activate the AutoML functionality. E.g, `self.naming = LassoRegression.name`. Further explaination is in section 2.2.
-#### 2.1.4 Define special_components
-Its purpose is to Invoke all special application functions for this algorithms by Scikit-learn framework.
-**Note:** The content of this part needs to be selected according to the actual situation of your own model.Can refer to similar classes.
-
-```
-GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
-```
-+ This line of code gets the image model output path from the environment variable.
-```
-GEOPI_OUTPUT_ARTIFACTS_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_PATH")
+(9) Define the instance attribute `customized` and `customized_name`
```
-+ This line of code takes the general output artifact path from the environment variable.
-**Note:** You need to choose to add the corresponding path according to the usage in the following functions.
-
-### 2.2 Add AutoML
-#### 2.2.1 Add AutoML code to class
-(1) Set AutoML related parameters
-```
- @property
- def settings(self) -> Dict:
- """The configuration of your model to implement AutoML by FLAML framework."""
- configuration = {
- "time_budget": '...'
- "metric": '...',
- "estimator_list": '...'
- "task": '...'
- }
- return configuration
+self.customized = True
+self.customized_name = "Algorithm Name"
```
-+ "time_budget" represents total running time in seconds
-+ "metric" represents Running metric
-+ "estimator_list" represents list of ML learners
-+ "task" represents task type
-**Note:** You can keep the parameters consistent, or you can modify them to make the AutoML better.
+These will be use to leverage the customization of AutlML functionality. E.g,`self.customized_name = "Lasso"`. Further explaination is in section 2.3.
-(2) Add parameters that need to be AutoML
-You can add the parameter tuning code according to the following code:
-```
- @property
- def customization(self) -> object:
- """The customized 'Your model' of FLAML framework."""
- from flaml import tune
- from flaml.data import 'TPYE'
- from flaml.model import SKLearnEstimator
- from sklearn.ensemble import 'model_name'
-
- class 'Model_Name'(SKLearnEstimator):
- def __init__(self, task=type, n_jobs=None, **config):
- super().__init__(task, **config)
- if task in 'TOYE':
- self.estimator_class = 'model_name'
-
- @classmethod
- def search_space(cls, data_size, task):
- space = {
- "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
- "'parameters2'": {"domain": tune.choice([True, False])},
- "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
- }
- return space
-
- return "Model_Name"
-```
-**Note1:** The content in ' ' needs to be modified according to your specific code
-**Note2:**
+(10) Define other instance attributes
```
- space = {
- "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
- "'parameters2'": {"domain": tune.choice([True, False])},
- "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
- }
+self.attributes=...
```
-+ tune.Uniform represents float
-+ tune.choice represents bool
-+ tune.randint represents int
-+ lower represents the minimum value of the range, upper represents the maximum value of the range, and init_value represents the initial value
-**Note:** You need to select parameters based on the actual situation of the model
-(3) Define special_components(FLAML)
-This part is the same as 2.1.4 as a whole, and can be modified with reference to it, but only the following two points need to be noted:
-a.The multi-dispatch function is different
-Scikit-learn framework:@dispatch()
-FLAML framework:@dispatch(bool)
+### 3.2 Add Manual Hyperparameter Tuning Functionality
-b.Added 'is_automl: bool' to the def
-**eg:**
-```
-Scikit-learn framework:
-def special_components(self, **kwargs) -> None:
+Our framework provides the user to set the algorithm hyperparameter manually or automiacally. In this part, we implement the manual functionality.
-FLAML framework:
-def special_components(self, is_automl: bool, **kwargs) -> None:
-```
-c.self.model has a different name
-**eg:**
-```
-Scikit-learn framework:
-coefficient=self.model.coefficient
-
-FLAML framework:
-coefficient=self.auto_model.coefficient
-```
+Sometimes the users want to input the hyperparameter values for model training manually, so you need to establish an interaction way to get the user's input.
-**Note:** You can refer to other similar codes to complete your code.
+#### 3.2.1 Define manual_hyper_parameters Method
-### 2.3 Get the hyperparameter value through interactive methods
-Sometimes the user wants to modify the hyperparameter values for model training, so you need to establish an interaction to get the user's modifications.
+The manual operation is control by the **manual_hyper_parameters** method. Inside this method, we encapsulate a lower level application function called algorithm_manual_hyper_parameters().
+```
+@classmethod
+def manual_hyper_parameters(cls) -> Dict:
+ """Manual hyper-parameters specification."""
+ print(f"-*-*- {cls.name} - Hyper-parameters Specification -*-*-")
+ hyper_parameters = algorithm_manual_hyper_parameters()
+ clear_output()
+ return hyper_parameters
+```
-#### 2.3.1 Find file
-You need to find the corresponding folder for model. The corresponding algorithm file is in the `func` folder in the model folder in the `data_mining` folder in the `geochemistrypi` folder.
++ The **manual_hyper_parameters** method is called in the corresponding mode operation file under the `geochemistrypi/data_mining/process` folder.
++ This lower level application function locates in the `geochemistrypi/data_mining/model/func/specific_mode` folder which limits the hyperparameters the user can set manually. E.g., If the model workflow class `LassoRegression` belongs to the regression mode, you need to add the `_lasso_regression.py` file under the folder `geochemistrypi/data_mining/model/func/algo_regression`. Here, `_lasso_regression.py` contains all encapsulated application functions specific to lasso algorithm.
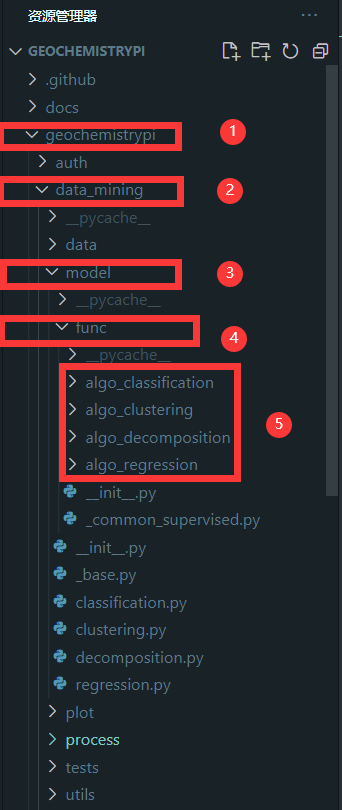
-**eg:** If your model belongs to the regression, you need to add the `corresponding.py` file in the `alog_regression` folder.
+#### 3.2.2 Create `_algorithm.py` File
+(1) Create a _algorithm.py file
-#### 2.3.2 Create the .py file and add content
-(1) Create a .py file
**Note:** Keep name format consistent.
(2) Import module
@@ -283,395 +237,644 @@ from ....constants import SECTION
```
from ....data.data_readiness import bool_input, float_input, num_input
```
-+ This needs to choose the appropriate import according to the hyperparameter type of model interaction.
++ You needs to choose the appropriate common utility functions according to the input type of hyperparameter.
-(3) Define the function
+(3) Define the application function
```
-def name_manual_hyper_parameters() -> Dict:
+def algorithm_manual_hyper_parameters() -> Dict:
```
-**Note:** The name needs to be consistent with that in 2.1.3.
(4) Interactive format
```
-print("Hyperparameters: Role")
-print("Recommended value")
-Hyperparameters = type_input(Recommended value, SECTION[2], "@Hyperparameters: ")
+print("Hyperparameters: Explaination")
+print("A good starting value ...")
+Hyperparameters = type_input(Default Value, SECTION[2], "@Hyperparameters: ")
```
-**Note:** The recommended value needs to be the default value of the corresponding package.
+**Note:** You can query ChatGPT for the recommended good starting value. The default value can come from that one in the imported library. For example, check the default value of the specific parameter for `Lasso` algorithm in [Scikit-learn Website](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html).
(5) Integrate all hyperparameters into a dictionary type and return.
```
hyper_parameters = {
- "Hyperparameters1": Hyperparameters1,
- "Hyperparameters": Hyperparameters2,
+ "Hyperparameters1": Hyperparameters1,
+ "Hyperparameters": Hyperparameters2,
}
retuen hyper_parameters
```
-#### 2.3.3 Import in the file that defines the model class
+#### 3.2.3 Import in The Model Workflow Class File
```
-from .func.algo_regression.Name import name_manual_hyper_parameters
+from .func.algo_mode._algorithm.py import algorithm_manual_hyper_parameters
```
-**eg:**

-### 2.4 Call Model
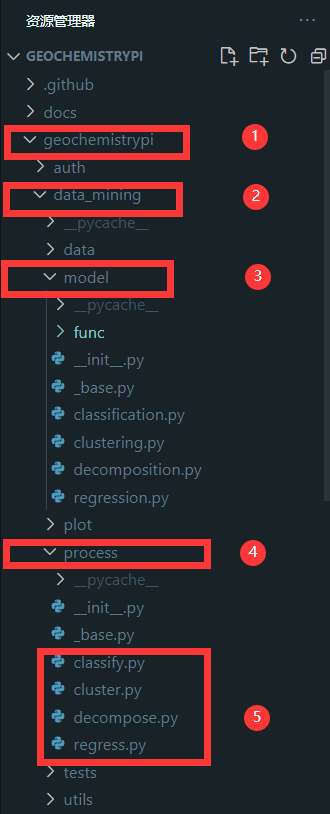
-#### 2.4.1 Find file
-Call the model in the corresponding file in the `process` folder. The corresponding algorithm file is in the `process` folder in the` model` folder in the `data_mining` folder in the `geochemistrypi` folder.
+### 3.3 Add Automated Hyperparameter Tuning (AutoML) Functionality
-
+#### 3.3.1 Add AutoML Code to Model Workflow Class
-**eg:** If your model belongs to the regression,you need to call it in the regress.py file.
+Currently, only supervised learning modes (regression and classification) support AutoML. Hence, only the algorithm belonging to these two modes need to implment AutoML functionality.
-#### 2.4.2 Import module
-You need to add your model in the from ..model.regression import().
-```
-from ..model.regression import(
- ...
- NAME,
-)
-```
-**Note:** NAME needs to be the same as the NAME when defining the class in step 2.1.2.
-**eg:**
+Our framework leverages FLAML + Ray to build the AutoML functionality. For some algorithms, FLAML has encapsulated them. Hence, it is easy to operate with those built-in algorithm. However, some algorithms without encapsulation needs our customization on our own.
-
+There are three cases in total:
++ C1: Encapsulated -> FLAML (Good example: `XGBoostRegression` in `regression.py`)
++ C2: Unencapsulated -> FLAML (Good example: `SVMRegression` in `regression.py`)
++ C3: Unencapsulated -> FLAML + RAY (Good example: `MLPRegression` in `regression.py`)
-#### 2.4.3 Call model
-There are two activate methods defined in the Regression and Classification algorithms, the first method uses the Scikit-learn framework, and the second method uses the FLAML and RAY frameworks. Decomposition and Clustering algorithms only use the Scikit-learn framework. Therefore, in the call, Regression and Classification need to add related codes to implement the call in both methods, and only one time is needed in Clustering and Decomposition.
+Here, we only talk about 2 cases, C1 and C2. C3 is a special case and it is only implemented in MLP algorithm.
-(1) Call model in the first activate method(Including Classification, Regression,Decomposition,Clustering)
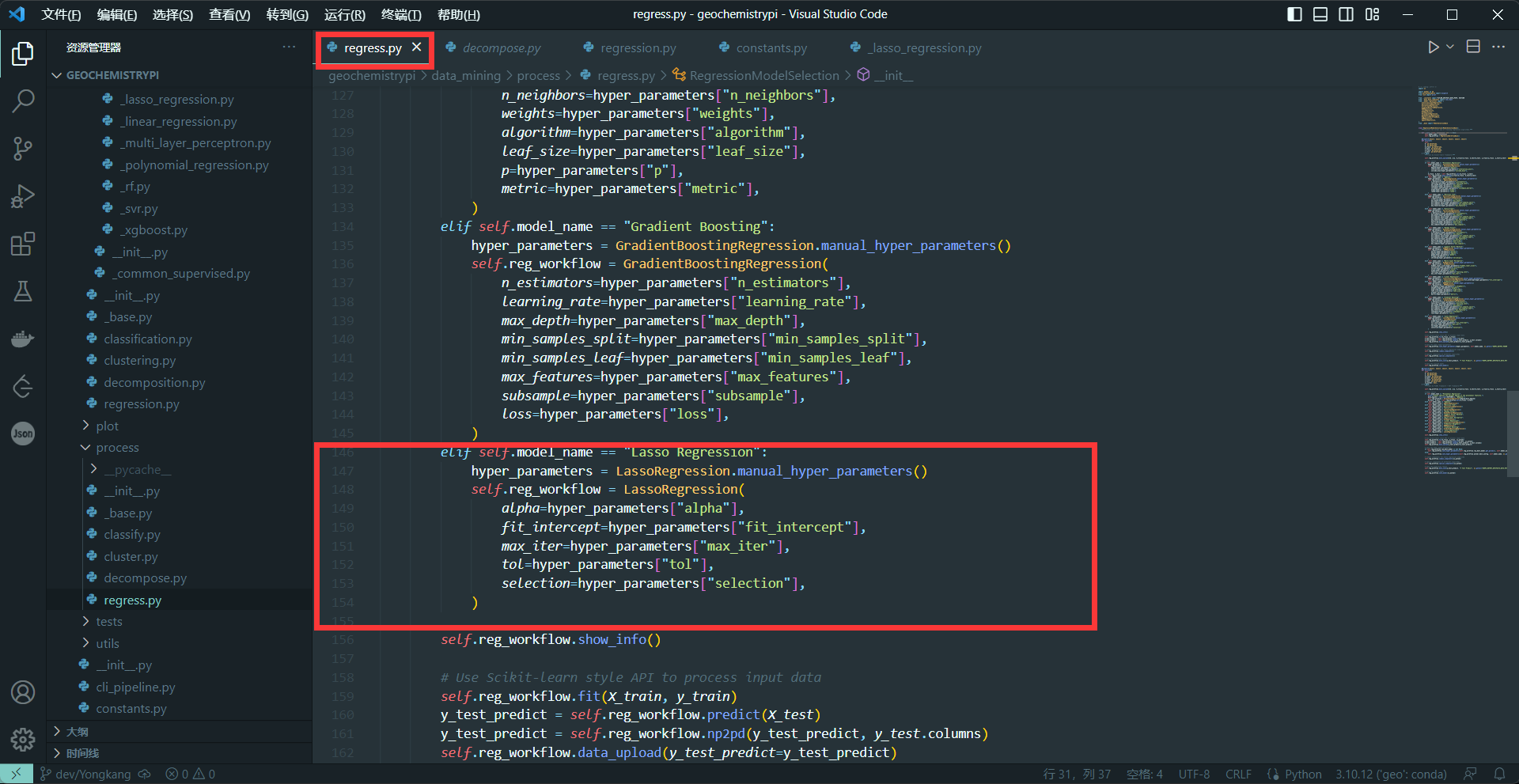
-```
-elif self.model_name == "name":
- hyper_parameters = NAME.manual_hyper_parameters()
- self.dcp_workflow = NAME(
- Hyperparameters1=hyper_parameters["Hyperparameters2"],
- Hyperparameters1=hyper_parameters["Hyperparameters2"],
- ...
- )
-```
-+ The name needs to be the same as the name in 2.4
-+ The hyperparameters in NAME() are the hyperparameters obtained interactively in 2.2
-**eg:**
-
+Noted:
-(2)Call model in the second activate method(Including Classification, Regression)
++ The calling method **fit** is defined in the base class, hence, no need to define it again in the specific model workflow class. You can refrence the **fit** method of `RegressionWorkflowBase` in `regression.py`
+
+The following two steps is needed to implement AutoML functionality in the model workflow class. But for C1 it only requires the first step while C2 needs two step both.
+
+(1) Create `settings` method
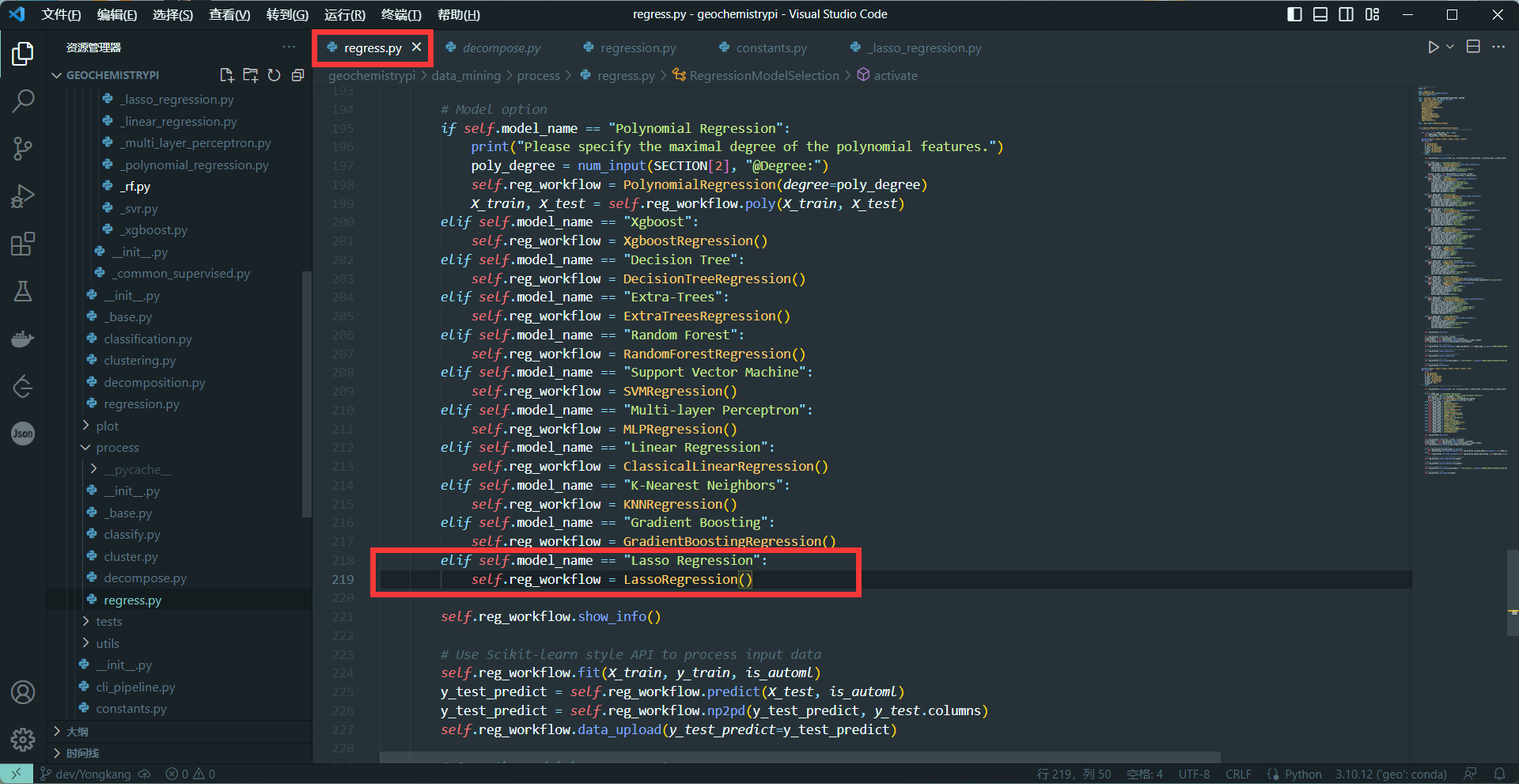
```
-elif self.model_name == "name":
- self.reg_workflow = NAME()
+@property
+def settings(self) -> Dict:
+ """The configuration of your model to implement AutoML by FLAML framework."""
+ configuration = {
+ "time_budget": '...'
+ "metric": '...',
+ "estimator_list": '...'
+ "task": '...'
+ }
+ return configuration
```
-+ The name needs to be the same as the name in 2.4
-**eg:**
-
++ "time_budget" represents total running time in seconds
++ "metric" represents Running metric
++ "estimator_list" represents list of ML learners
++ "task" represents task type
-### 2.5 Add the algorithm list and set NON_AUTOML_MODELS
+For C1, the value of "estimator_list" should come from the specified name in [FLAML library](https://microsoft.github.io/FLAML/docs/Use-Cases/Task-Oriented-AutoML). For example, the specified name `xgboost` in the model workflow class `XGBoostRegression`. Also we need to put this specified value inside a list.
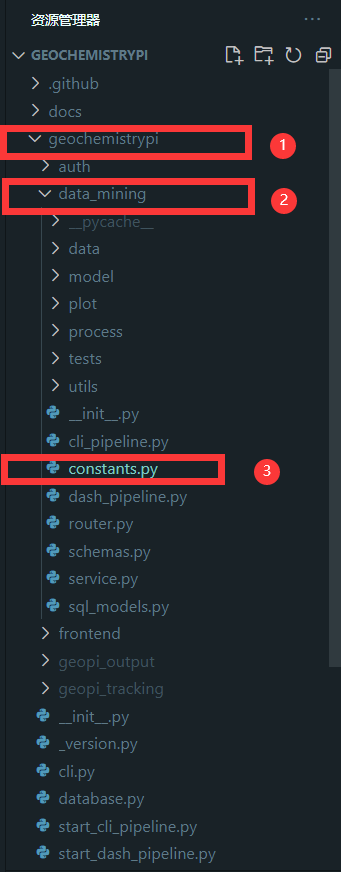
-#### 2.5.1 Find file
-Find the constants file to add the model name,The constants file is in the `data_mining` folder in the `geochemistrypi` folder.
+
- - [2.5 Add the algorithm list and set NON\_AUTOML\_MODELS](#25-add-the-algorithm-list-and-set-non_automl_models)
- - [2.5.1 Find file](#251-find-file)
+The code of each layer lies as shown above.
- - [2.6 Add Functionality](#26-add-functionality)
+**Notice**: in our framework, a **model workflow class** refers to an **algorithm workflow class** and a **mode** includes multiple model workflow classes.
- - [2.6.1 Model Research](#261-model-research)
+Now, we will take KMeans algorithm as an example to illustrate the connection between each layer. Don't get too hung up on ths part. Once you finish reading the whole article, you can come back to here again.
- - [2.6.2 Add Common_component](#262-add-common_component)
+After reading this article, you are recommended to refer to this publication also for more details on the whole scope of our framework:
- - [2.6.3 Add Special_component](#263-add-special_component)
+https://agupubs.onlinelibrary.wiley.com/doi/10.1029/2023GC011324
-- [3. Test model](#3-test-model)
-- [4. Completed Pull Request](#4-completed-pull-request)
+## 2. Understand Machine Learning Algorithm
+You need to understand the general meaning of the machine learning algorithm you are responsible for. Then you encapsultate it as an algorithm workflow in our framework and put it under the directory `geochemistrypi/data_mining/model`. Then you need to determine which **mode** this algorithm belongs to and the role of each parameter. For example, linear regression algorithm belongs to regression mode in our framework.
-- [5. Precautions](#5-precautions)
++ When learning the ML algorithm, you can refer to the relevant knowledge on the [scikit-learn official website](https://scikit-learn.org/stable/index.html).
-## 1. Understand the model
-You need to understand the general meaning of the model, determine which algorithm the model belongs to and the role of each parameter.
-+ You can choose to learn about the relevant knowledge on the [scikit-learn official website](https://scikit-learn.org/stable/index.html).
+## 3. Construct Model Workflow Class
+**Noted**: You can reference any existing model workflow classes in our framework to implement your own model workflow class.
-## 2. Add Model
-### 2.1 Add The Model Class
-#### 2.1.1 Find Add File
-First, you need to define the model class that you need to complete in the corresponding algorithm file. The corresponding algorithm file is in the `model` folder in the `data_mining` folder in the `geochemistrypi` folder.
+### 3.1 Add Basic Elements
+
+#### 3.1.1 Find File
+First, you need to construct the algorithm workflow class in the corresponding model file. The corresponding model file locates under the path `geochemistrypi/data_mining/model`.

-**eg:** If you want to add a model to the regression algorithm, you need to add it in the `regression.py` file.
+**E.g.,** If you want to add a model for the regression mode, you need to add it in the `regression.py` file.
+
+#### 3.1.2 Define Class Attributes and Constructor
+
+(1) Define the algorithm workflow class and its base class
-#### 2.1.2 Define class properties and constructors, etc.
-(1) Define the class and the required Base class
```
-class NAME (Base class):
+class ModelWorkflowClassName(BaseModelWorkflowClassName):
```
-+ NAME is the name of the algorithm, you can refer to the NAME of other models, the format needs to be consistent.
-+ Base class needs to be selected according to the actual algorithm requirements.
++ You can refer to the ModelName of other models, the format (Upper case and with the suffix 'Corresponding Mode') needs to be consistent. E.g., `XGBoostRegression`.
++ Base class needs to be inherited according to the mode the model belongs to.
```
-"""The automation workflow of using "Name" to make insightful products."""
+"""The automation workflow of using "ModelWorkflowClassName" algorithm to make insightful products."""
```
-+ Class explanation, you can refer to other classes.
++ Class docstring, you can refer to other classes. The template is shown above.
-(2) Define the name and the special_function
+(2) Define the class attributes `name`
```
-name = "name"
+name = "algorithm terminology"
```
-+ Define name, different from NMAE.
-+ This name needs to be added to the _`constants.py`_ file and the corresponding algorithm file in the `process` folder. Note that the names are consistent.
++ The class attributes `name` is different from ModelWorkflowClassName. E.g., the name `XGBoost` in `XGBoostRegression` model workflow class.
++ This name needs to be added to the corresponding constant variable in `geochemistrypi/data_mining/constants.py` file and the corresponding mode processing file under the `geochemistrypi/data_mining/process` folder. Note that those name value should be identical. It will be further explained in later section.
++ For example, the name value `XGBoost` should be included in the constant varible `REGRESSION_MODELS` in `geochemistrypi/data_mining/constants.py` file and it will be use in `geochemistrypi/data_mining/process/regress.py`.
+
+(3) Define the class attrbutes `common_functiion` or `special_function`
+
+If this model workflow class is a base class, you need to define the class attrbutes `common_functiion`. For example, the class attrbutes `common_functiion` in the base workflow class `RegressionWorkflowBase`.
+
+The values of `common_functiion` are the description of the functionalities of the models belonging to the same mode. It means the children class (all regession models) can share the same common functionalies as well.
+
+```
+common_functiion = []
+```
+
+If this model workflow class is a specific model workflow class, you need to define the class attrbutes `special_function`. For example, the class attrbutes `special_function` in the model workflow class `XGBoostRegression`.
+
+The values of `special_function` are the description of the owned functionalities of that specific model. Those special functions cannot be reused by other models.
+
```
special_function = []
```
-+ special_function is added according to the specific situation of the model, you can refer to other similar models.
-(3) Define constructor
+More detail will be explained in the later section.
+
+(4) Define the signature of the constructor
```
def __init__(
self,
- parameter:type=Default parameter value,
+ parameter: type = Default parameter value,
) -> None:
```
-+ All parameters in the corresponding model function need to be written out.
-+ Default parameter value needs to be set according to official documents.
++ The parameters in the constructor is from the algorithm library you depend on. For example, you use **Lasso** algorithm from Sckit-learn library. You can reference its introduction ([Lasso](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html)) in Scikit-learn website.
++ Default parameter value needs to be set according to scikit-learn official documents also.
+
+
+
```
- """
+"""
Parameters
----------
-parameter:type,default = Dedault
+parameter: type,default = Dedault
References
----------
Scikit-learn API: sklearn.model.name
https://scikit-learn.org/......
```
-+ Parameters is in the source of the corresponding model on the official website of sklearn
-
-**eg:** Take the [Lasso algorithm](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html) as a column.
-
-
++ Parameters docstring are in the source code of the corresponding algorithm on the official website of sklearn.

-+ References is your model's official website.
-(4) The constructor of Base class is called
+(5) The constructor of Base class is called
```
super().__init__()
```
-(5) Initializes the instance's state by assigning the parameter values passed to the constructor to the instance's properties.
+
+(6) Initializes the instance's state by assigning the parameter values passed to the constructor to the instance's attributes.
```
self.parameter=parameter
```
-(6) Create the model and assign
+
+(7) Instantiate the algorithm class you depend on and assign. For example, `Lasso` from the library `sklearn.linear_model`.
```
-self.model=modelname(
+self.model = modelname(
parameter=self.parameter
)
```
-**Note:** Don't forget to import Model from scikit-learn
+**Note:** Don't forget to import the class from scikit-learn library

-(7) Define other class properties
+(8) Define the instance attribute `naming`
```
-self.properties=...
-```
-
-#### 2.1.3 Define manual_hyper_parameters
-manual_hyper_parameters gets the hyperparameter value by calling the manual hyperparameter function, and returns hyper_parameters.
+self.naming = Class.name
```
-hyper_parameters = name_manual_hyper_parameters()
-```
-+ This function calls the corresponding function in the `func` folder (needs to be written, see 2.2.2) to get the hyperparameter value.
-
-+ This function is called in the corresponding file of the `Process` folder (need to be written, see 2.3).
-+ Can be written with reference to similar classes
-
+This one will be use to print the name of the class and to activate the AutoML functionality. E.g, `self.naming = LassoRegression.name`. Further explaination is in section 2.2.
-#### 2.1.4 Define special_components
-Its purpose is to Invoke all special application functions for this algorithms by Scikit-learn framework.
-**Note:** The content of this part needs to be selected according to the actual situation of your own model.Can refer to similar classes.
-
-```
-GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
-```
-+ This line of code gets the image model output path from the environment variable.
-```
-GEOPI_OUTPUT_ARTIFACTS_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_PATH")
+(9) Define the instance attribute `customized` and `customized_name`
```
-+ This line of code takes the general output artifact path from the environment variable.
-**Note:** You need to choose to add the corresponding path according to the usage in the following functions.
-
-### 2.2 Add AutoML
-#### 2.2.1 Add AutoML code to class
-(1) Set AutoML related parameters
-```
- @property
- def settings(self) -> Dict:
- """The configuration of your model to implement AutoML by FLAML framework."""
- configuration = {
- "time_budget": '...'
- "metric": '...',
- "estimator_list": '...'
- "task": '...'
- }
- return configuration
+self.customized = True
+self.customized_name = "Algorithm Name"
```
-+ "time_budget" represents total running time in seconds
-+ "metric" represents Running metric
-+ "estimator_list" represents list of ML learners
-+ "task" represents task type
-**Note:** You can keep the parameters consistent, or you can modify them to make the AutoML better.
+These will be use to leverage the customization of AutlML functionality. E.g,`self.customized_name = "Lasso"`. Further explaination is in section 2.3.
-(2) Add parameters that need to be AutoML
-You can add the parameter tuning code according to the following code:
-```
- @property
- def customization(self) -> object:
- """The customized 'Your model' of FLAML framework."""
- from flaml import tune
- from flaml.data import 'TPYE'
- from flaml.model import SKLearnEstimator
- from sklearn.ensemble import 'model_name'
-
- class 'Model_Name'(SKLearnEstimator):
- def __init__(self, task=type, n_jobs=None, **config):
- super().__init__(task, **config)
- if task in 'TOYE':
- self.estimator_class = 'model_name'
-
- @classmethod
- def search_space(cls, data_size, task):
- space = {
- "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
- "'parameters2'": {"domain": tune.choice([True, False])},
- "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
- }
- return space
-
- return "Model_Name"
-```
-**Note1:** The content in ' ' needs to be modified according to your specific code
-**Note2:**
+(10) Define other instance attributes
```
- space = {
- "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
- "'parameters2'": {"domain": tune.choice([True, False])},
- "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
- }
+self.attributes=...
```
-+ tune.Uniform represents float
-+ tune.choice represents bool
-+ tune.randint represents int
-+ lower represents the minimum value of the range, upper represents the maximum value of the range, and init_value represents the initial value
-**Note:** You need to select parameters based on the actual situation of the model
-(3) Define special_components(FLAML)
-This part is the same as 2.1.4 as a whole, and can be modified with reference to it, but only the following two points need to be noted:
-a.The multi-dispatch function is different
-Scikit-learn framework:@dispatch()
-FLAML framework:@dispatch(bool)
+### 3.2 Add Manual Hyperparameter Tuning Functionality
-b.Added 'is_automl: bool' to the def
-**eg:**
-```
-Scikit-learn framework:
-def special_components(self, **kwargs) -> None:
+Our framework provides the user to set the algorithm hyperparameter manually or automiacally. In this part, we implement the manual functionality.
-FLAML framework:
-def special_components(self, is_automl: bool, **kwargs) -> None:
-```
-c.self.model has a different name
-**eg:**
-```
-Scikit-learn framework:
-coefficient=self.model.coefficient
-
-FLAML framework:
-coefficient=self.auto_model.coefficient
-```
+Sometimes the users want to input the hyperparameter values for model training manually, so you need to establish an interaction way to get the user's input.
-**Note:** You can refer to other similar codes to complete your code.
+#### 3.2.1 Define manual_hyper_parameters Method
-### 2.3 Get the hyperparameter value through interactive methods
-Sometimes the user wants to modify the hyperparameter values for model training, so you need to establish an interaction to get the user's modifications.
+The manual operation is control by the **manual_hyper_parameters** method. Inside this method, we encapsulate a lower level application function called algorithm_manual_hyper_parameters().
+```
+@classmethod
+def manual_hyper_parameters(cls) -> Dict:
+ """Manual hyper-parameters specification."""
+ print(f"-*-*- {cls.name} - Hyper-parameters Specification -*-*-")
+ hyper_parameters = algorithm_manual_hyper_parameters()
+ clear_output()
+ return hyper_parameters
+```
-#### 2.3.1 Find file
-You need to find the corresponding folder for model. The corresponding algorithm file is in the `func` folder in the model folder in the `data_mining` folder in the `geochemistrypi` folder.
++ The **manual_hyper_parameters** method is called in the corresponding mode operation file under the `geochemistrypi/data_mining/process` folder.
++ This lower level application function locates in the `geochemistrypi/data_mining/model/func/specific_mode` folder which limits the hyperparameters the user can set manually. E.g., If the model workflow class `LassoRegression` belongs to the regression mode, you need to add the `_lasso_regression.py` file under the folder `geochemistrypi/data_mining/model/func/algo_regression`. Here, `_lasso_regression.py` contains all encapsulated application functions specific to lasso algorithm.

-**eg:** If your model belongs to the regression, you need to add the `corresponding.py` file in the `alog_regression` folder.
+#### 3.2.2 Create `_algorithm.py` File
+(1) Create a _algorithm.py file
-#### 2.3.2 Create the .py file and add content
-(1) Create a .py file
**Note:** Keep name format consistent.
(2) Import module
@@ -283,395 +237,644 @@ from ....constants import SECTION
```
from ....data.data_readiness import bool_input, float_input, num_input
```
-+ This needs to choose the appropriate import according to the hyperparameter type of model interaction.
++ You needs to choose the appropriate common utility functions according to the input type of hyperparameter.
-(3) Define the function
+(3) Define the application function
```
-def name_manual_hyper_parameters() -> Dict:
+def algorithm_manual_hyper_parameters() -> Dict:
```
-**Note:** The name needs to be consistent with that in 2.1.3.
(4) Interactive format
```
-print("Hyperparameters: Role")
-print("Recommended value")
-Hyperparameters = type_input(Recommended value, SECTION[2], "@Hyperparameters: ")
+print("Hyperparameters: Explaination")
+print("A good starting value ...")
+Hyperparameters = type_input(Default Value, SECTION[2], "@Hyperparameters: ")
```
-**Note:** The recommended value needs to be the default value of the corresponding package.
+**Note:** You can query ChatGPT for the recommended good starting value. The default value can come from that one in the imported library. For example, check the default value of the specific parameter for `Lasso` algorithm in [Scikit-learn Website](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html).
(5) Integrate all hyperparameters into a dictionary type and return.
```
hyper_parameters = {
- "Hyperparameters1": Hyperparameters1,
- "Hyperparameters": Hyperparameters2,
+ "Hyperparameters1": Hyperparameters1,
+ "Hyperparameters": Hyperparameters2,
}
retuen hyper_parameters
```
-#### 2.3.3 Import in the file that defines the model class
+#### 3.2.3 Import in The Model Workflow Class File
```
-from .func.algo_regression.Name import name_manual_hyper_parameters
+from .func.algo_mode._algorithm.py import algorithm_manual_hyper_parameters
```
-**eg:**

-### 2.4 Call Model
-#### 2.4.1 Find file
-Call the model in the corresponding file in the `process` folder. The corresponding algorithm file is in the `process` folder in the` model` folder in the `data_mining` folder in the `geochemistrypi` folder.
+### 3.3 Add Automated Hyperparameter Tuning (AutoML) Functionality
-
+#### 3.3.1 Add AutoML Code to Model Workflow Class
-**eg:** If your model belongs to the regression,you need to call it in the regress.py file.
+Currently, only supervised learning modes (regression and classification) support AutoML. Hence, only the algorithm belonging to these two modes need to implment AutoML functionality.
-#### 2.4.2 Import module
-You need to add your model in the from ..model.regression import().
-```
-from ..model.regression import(
- ...
- NAME,
-)
-```
-**Note:** NAME needs to be the same as the NAME when defining the class in step 2.1.2.
-**eg:**
+Our framework leverages FLAML + Ray to build the AutoML functionality. For some algorithms, FLAML has encapsulated them. Hence, it is easy to operate with those built-in algorithm. However, some algorithms without encapsulation needs our customization on our own.
-
+There are three cases in total:
++ C1: Encapsulated -> FLAML (Good example: `XGBoostRegression` in `regression.py`)
++ C2: Unencapsulated -> FLAML (Good example: `SVMRegression` in `regression.py`)
++ C3: Unencapsulated -> FLAML + RAY (Good example: `MLPRegression` in `regression.py`)
-#### 2.4.3 Call model
-There are two activate methods defined in the Regression and Classification algorithms, the first method uses the Scikit-learn framework, and the second method uses the FLAML and RAY frameworks. Decomposition and Clustering algorithms only use the Scikit-learn framework. Therefore, in the call, Regression and Classification need to add related codes to implement the call in both methods, and only one time is needed in Clustering and Decomposition.
+Here, we only talk about 2 cases, C1 and C2. C3 is a special case and it is only implemented in MLP algorithm.
-(1) Call model in the first activate method(Including Classification, Regression,Decomposition,Clustering)
-```
-elif self.model_name == "name":
- hyper_parameters = NAME.manual_hyper_parameters()
- self.dcp_workflow = NAME(
- Hyperparameters1=hyper_parameters["Hyperparameters2"],
- Hyperparameters1=hyper_parameters["Hyperparameters2"],
- ...
- )
-```
-+ The name needs to be the same as the name in 2.4
-+ The hyperparameters in NAME() are the hyperparameters obtained interactively in 2.2
-**eg:**
-
+Noted:
-(2)Call model in the second activate method(Including Classification, Regression)
++ The calling method **fit** is defined in the base class, hence, no need to define it again in the specific model workflow class. You can refrence the **fit** method of `RegressionWorkflowBase` in `regression.py`
+
+The following two steps is needed to implement AutoML functionality in the model workflow class. But for C1 it only requires the first step while C2 needs two step both.
+
+(1) Create `settings` method
```
-elif self.model_name == "name":
- self.reg_workflow = NAME()
+@property
+def settings(self) -> Dict:
+ """The configuration of your model to implement AutoML by FLAML framework."""
+ configuration = {
+ "time_budget": '...'
+ "metric": '...',
+ "estimator_list": '...'
+ "task": '...'
+ }
+ return configuration
```
-+ The name needs to be the same as the name in 2.4
-**eg:**
-
++ "time_budget" represents total running time in seconds
++ "metric" represents Running metric
++ "estimator_list" represents list of ML learners
++ "task" represents task type
-### 2.5 Add the algorithm list and set NON_AUTOML_MODELS
+For C1, the value of "estimator_list" should come from the specified name in [FLAML library](https://microsoft.github.io/FLAML/docs/Use-Cases/Task-Oriented-AutoML). For example, the specified name `xgboost` in the model workflow class `XGBoostRegression`. Also we need to put this specified value inside a list.
-#### 2.5.1 Find file
-Find the constants file to add the model name,The constants file is in the `data_mining` folder in the `geochemistrypi` folder.
+ -
+For C2, the value of "estimator_list" should be the instance attribute `self.customized_name`. For example, `self.customized_name = "SVR"` in the model workflow class `SVMRegression`. Also we need to put this specified value inside a list.
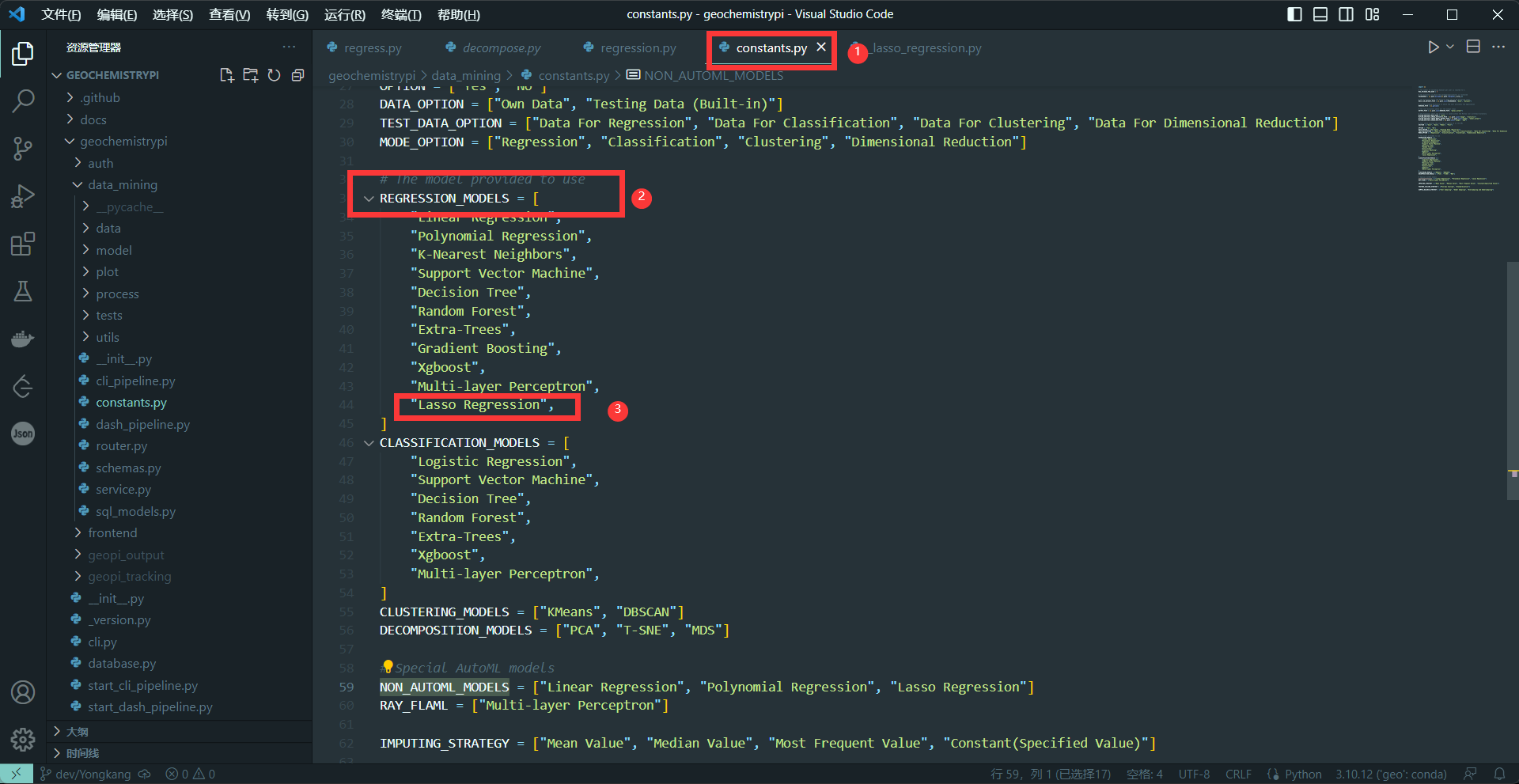
-(1) Add the model name
-Add model name to the algorithm list corresponding to the model in the constants file.
-**eg:** Add the name of the Lasso regression algorithm.
-
-
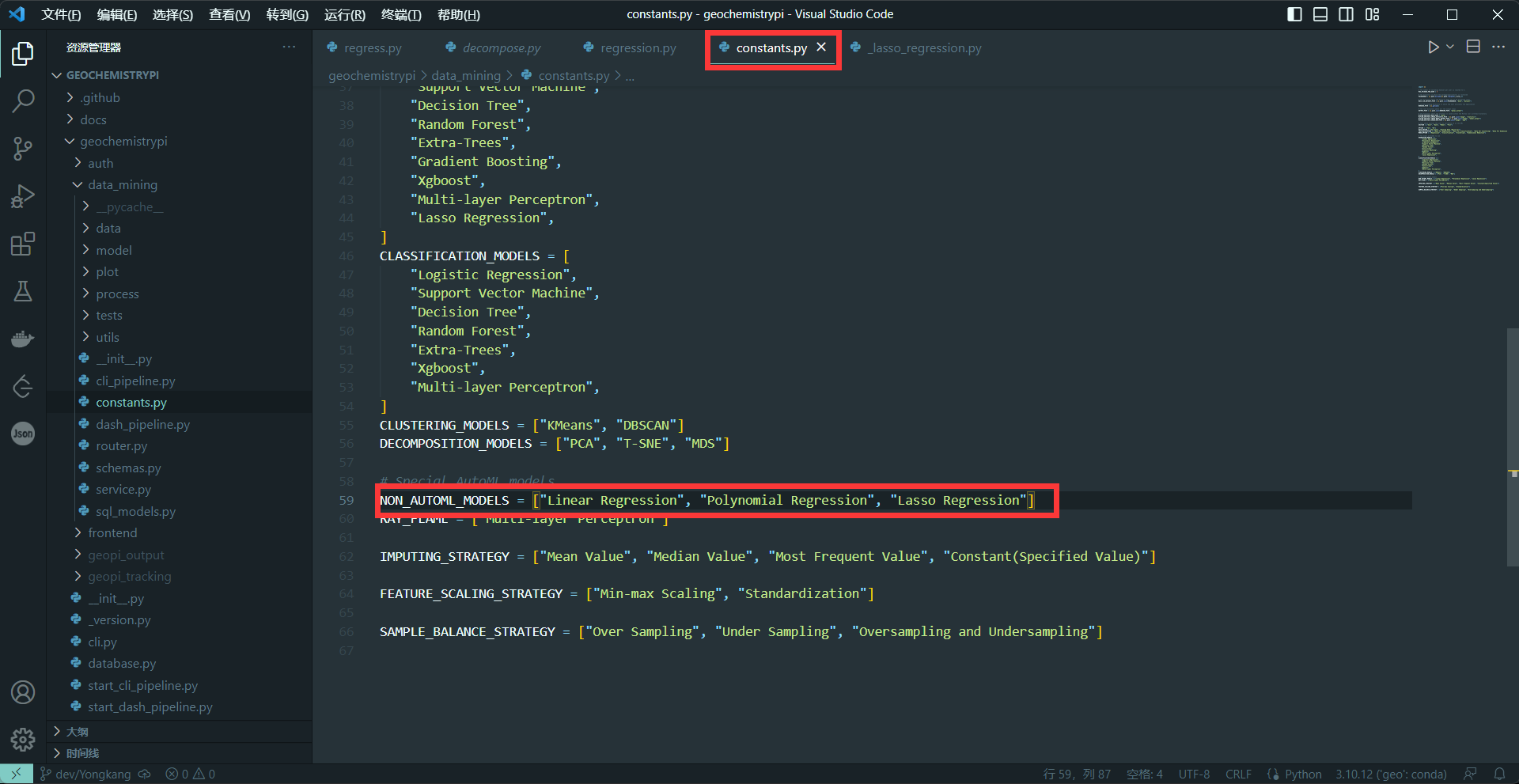
-(2)set NON_AUTOML_MODELS
-Because this is a tutorial without automatic parameters, you need to add the model name in the NON_AUTOML_MODELS.
-**eg:**
-
+**Note:** You can keep the other key-value pair consistent with other exited model workflow classes.
+(2) Create `customization` method
+You can add the parameter tuning code according to the following code:
+```
+@property
+def customization(self) -> object:
+ """The customized 'Your model' of FLAML framework."""
+ from flaml import tune
+ from flaml.data import 'TPYE'
+ from flaml.model import SKLearnEstimator
+ from 'sklearn' import 'model_name'
+
+ class 'Model_Name'(SKLearnEstimator):
+ def __init__(self, task=type, n_jobs=None, **config):
+ super().__init__(task, **config)
+ if task in 'TYPE':
+ self.estimator_class = 'model_name'
+
+ @classmethod
+ def search_space(cls, data_size, task):
+ space = {
+ "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
+ "'parameters2'": {"domain": tune.choice([True, False])},
+ "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
+ }
+ return space
+
+ return "Model_Name"
+```
+**Note1:** The content in ' ' needs to be modified according to your specific code. You can reference that one in the model workflow class `SVMRegression`.
+**Note2:**
+```
+space = {
+ "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
+ "'parameters2'": {"domain": tune.choice([True, False])},
+ "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
+}
+```
++ tune.uniform represents float
++ tune.choice represents bool
++ tune.randint represents int
++ lower represents the minimum value of the range, upper represents the maximum value of the range, and init_value represents the initial value
+**Note:** You need to select parameters based on the actual situation of the model
+### 3.4 Add Application Function to Model Workflow Class
-### 2.6 Add Functionality
+We treat the insightful outputs (index, scores) or diagrams to help to analyze and understand the algorithm as useful application. For example, XGBoost algorithm can produce feature importance score, hence, drawing feature importance diagram is an **application function** we can add to the model workflow class `XGBoostRegression`.
-#### 2.6.1 Model Research
+Conduct research on the corresponding model and look for its useful application functions that need to be added.
-Conduct research on the corresponding model and confirm the functions that need to be added.
++ You can confirm the functions that need to be added on the official website of the model (such as scikit learn), search engines (such as Google), chatGPT, etc.
-\+ You can confirm the functions that need to be added on the official website of the model (such as scikit learn), search engines (such as Google), chatGPT, etc.
+In our framework, we define two types of application function: **common application function** and **special application function**.
-(1) Common_component is a public function in a class, and all functions in each class can be used, so they need to be added in the parent class,Each of the parent classes can call Common_component.
+Common application function can be shared among the model workflow classes which belong to the same mode. It will be placed inside the base model workflow class. For example, `classification_report` is a common application function placed inside the base class `ClassificationWorkflowBase`. Notice that it is encapsulated in the **private** instance method `_classification_report`.
-(2) Special_component is unique to the model, so they need to be added in a specific model,Only they can use it.
+Likewise, special application function is the special fucntionalities owned by the algorithm itself, hence it is placed inside a specific model workflow class. For example, for KMeans algorithm, we can get the inertia scores from it. Hence, inside the model workflow class `KMeansClustering`, we have a **private** instance method `_get_inertia_scores`.
-
+Now, the next question is how to invoke these application function in our framework.
+In fact, we put the invocation of the application function in the component method. Accordingly, we have two types of components:
+(1) `common_components` is a public method in the base class, and all common application functions will be invoked inside.
-#### 2.6.2 Add Common_component
+(2) `special_components` is unique to the algorithm, so they need to be added in a specific model workflow class. All special aaplication function related to this algorithm will be invoked inside.
-Common_component refer to functions that can be used by all internal submodels, so it is necessary to consider the situation of each submodel when adding them.
+
-***\*1. Add corresponding functionality to the parent class\****
+For more details, you can refer to the brief illustraion of the framework in section 1.
-Once you've identified the features you want to add, you can define the corresponding functions in the parent class.
+#### 3.4.1 Add Common Application Functions and `common_components` Method
-The code format is:
+`common_components` will invoke the common application functions used by all its children model workflow class, so it is necessary to consider the situation of each child model workflow class when adding a application function to it. The better way is to put the application function inside a specific child model workflow class firstly if you are not sure it can be classified as a common application function.
-(1) Define the function name and add the required parameters.
+**1. Add common application function to the base class**
-(2) Use annotations to describe function functionsUse annotations to describe function functions.
+Once you’ve identified the functionality you want to add, you can define the corresponding functions in the base class.
-(3) Referencing specific functions to implement functionality.
+The steps to implement are:
-(4) Change the format of data acquisition and save data or images.
+(1) Define the private function name and add the required parameters.
+(2) Use annotations to decorate the function.
+(3) Add the docstring to explain the use of this functionality.
+(4) Referencing specific libraries (e.g., Scikit-learn) to implement the functionality.
+(5) Change the format of data acquisition and save the produced data or images, etc.
+**2. Encapsulte the concrete code in Layer 1**
+Please refer to our framework's definition of **Layer 1** in section 1.
-***\*2. Define Common_component\****
+Some functions may use large code due to their complexity. To ensure the style and readability of the codebase, you need to put the specific function implementation into the corresponding `geochemistrypi/data_mining/model/func/mode/_common` files and call it.
-(1) Define the common_components in the parent class, its role is to set where the output is saved.
+The steps to implement are:
-(2) Set the parameter source for the added function.
+(1) Define the public function name, add the required parameters and proper decorator.
+(2) Add the docstring to explain the use of this functionality,the significance of each parameter and the related reference.
+(3) Implement functionality.
+(4) Returns the value used in **Layer 2**.
+**3. Define `common_components` Method**
+The steps to implement are:
-***\*3. Implement function functions\****
+(1) Define the path to store the data and images, etc.
+(2) Invoke the common application functions one by one.
-Some functions may use large code due to their complexity. To ensure the style and readability of the code, you need to put the specific function implementation into the corresponding `_common` files and call it.
+**4. Apeend The Name of Functionality in Class Attribute `common_function`**
-It includes:
+The steps to implement are:
-(1) Explain the significance of each parameter.
+(1) Create a class attribute `common_function` list in `ClusteringWorkflowBase`
+(2) Create a enum class to include the name of the functionality
+(3) Append the value of enum class into `common_function` list
-(2) Implement functionality.
+**Example**
-(3) Returns the required parameters.
+The following is the example of adding model evaluation score to the clustering base class.
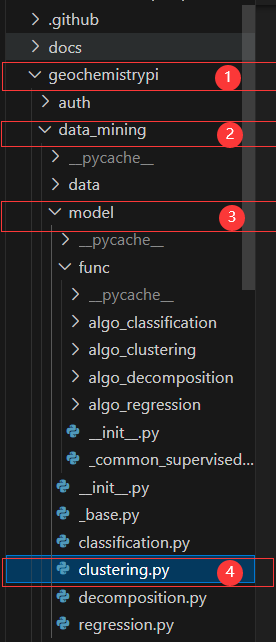
+First, you need to find the base class of clustering.
+
-***\*eg:\**** You want to add model evaluation to your clustering.
+
-
+For C2, the value of "estimator_list" should be the instance attribute `self.customized_name`. For example, `self.customized_name = "SVR"` in the model workflow class `SVMRegression`. Also we need to put this specified value inside a list.
-(1) Add the model name
-Add model name to the algorithm list corresponding to the model in the constants file.
-**eg:** Add the name of the Lasso regression algorithm.
-
-
-(2)set NON_AUTOML_MODELS
-Because this is a tutorial without automatic parameters, you need to add the model name in the NON_AUTOML_MODELS.
-**eg:**
-
+**Note:** You can keep the other key-value pair consistent with other exited model workflow classes.
+(2) Create `customization` method
+You can add the parameter tuning code according to the following code:
+```
+@property
+def customization(self) -> object:
+ """The customized 'Your model' of FLAML framework."""
+ from flaml import tune
+ from flaml.data import 'TPYE'
+ from flaml.model import SKLearnEstimator
+ from 'sklearn' import 'model_name'
+
+ class 'Model_Name'(SKLearnEstimator):
+ def __init__(self, task=type, n_jobs=None, **config):
+ super().__init__(task, **config)
+ if task in 'TYPE':
+ self.estimator_class = 'model_name'
+
+ @classmethod
+ def search_space(cls, data_size, task):
+ space = {
+ "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
+ "'parameters2'": {"domain": tune.choice([True, False])},
+ "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
+ }
+ return space
+
+ return "Model_Name"
+```
+**Note1:** The content in ' ' needs to be modified according to your specific code. You can reference that one in the model workflow class `SVMRegression`.
+**Note2:**
+```
+space = {
+ "'parameters1'": {"domain": tune.uniform(lower='...', upper='...'), "init_value": '...'},
+ "'parameters2'": {"domain": tune.choice([True, False])},
+ "'parameters3'": {"domain": tune.randint(lower='...', upper='...'), "init_value": '...'},
+}
+```
++ tune.uniform represents float
++ tune.choice represents bool
++ tune.randint represents int
++ lower represents the minimum value of the range, upper represents the maximum value of the range, and init_value represents the initial value
+**Note:** You need to select parameters based on the actual situation of the model
+### 3.4 Add Application Function to Model Workflow Class
-### 2.6 Add Functionality
+We treat the insightful outputs (index, scores) or diagrams to help to analyze and understand the algorithm as useful application. For example, XGBoost algorithm can produce feature importance score, hence, drawing feature importance diagram is an **application function** we can add to the model workflow class `XGBoostRegression`.
-#### 2.6.1 Model Research
+Conduct research on the corresponding model and look for its useful application functions that need to be added.
-Conduct research on the corresponding model and confirm the functions that need to be added.
++ You can confirm the functions that need to be added on the official website of the model (such as scikit learn), search engines (such as Google), chatGPT, etc.
-\+ You can confirm the functions that need to be added on the official website of the model (such as scikit learn), search engines (such as Google), chatGPT, etc.
+In our framework, we define two types of application function: **common application function** and **special application function**.
-(1) Common_component is a public function in a class, and all functions in each class can be used, so they need to be added in the parent class,Each of the parent classes can call Common_component.
+Common application function can be shared among the model workflow classes which belong to the same mode. It will be placed inside the base model workflow class. For example, `classification_report` is a common application function placed inside the base class `ClassificationWorkflowBase`. Notice that it is encapsulated in the **private** instance method `_classification_report`.
-(2) Special_component is unique to the model, so they need to be added in a specific model,Only they can use it.
+Likewise, special application function is the special fucntionalities owned by the algorithm itself, hence it is placed inside a specific model workflow class. For example, for KMeans algorithm, we can get the inertia scores from it. Hence, inside the model workflow class `KMeansClustering`, we have a **private** instance method `_get_inertia_scores`.
-
+Now, the next question is how to invoke these application function in our framework.
+In fact, we put the invocation of the application function in the component method. Accordingly, we have two types of components:
+(1) `common_components` is a public method in the base class, and all common application functions will be invoked inside.
-#### 2.6.2 Add Common_component
+(2) `special_components` is unique to the algorithm, so they need to be added in a specific model workflow class. All special aaplication function related to this algorithm will be invoked inside.
-Common_component refer to functions that can be used by all internal submodels, so it is necessary to consider the situation of each submodel when adding them.
+
-***\*1. Add corresponding functionality to the parent class\****
+For more details, you can refer to the brief illustraion of the framework in section 1.
-Once you've identified the features you want to add, you can define the corresponding functions in the parent class.
+#### 3.4.1 Add Common Application Functions and `common_components` Method
-The code format is:
+`common_components` will invoke the common application functions used by all its children model workflow class, so it is necessary to consider the situation of each child model workflow class when adding a application function to it. The better way is to put the application function inside a specific child model workflow class firstly if you are not sure it can be classified as a common application function.
-(1) Define the function name and add the required parameters.
+**1. Add common application function to the base class**
-(2) Use annotations to describe function functionsUse annotations to describe function functions.
+Once you’ve identified the functionality you want to add, you can define the corresponding functions in the base class.
-(3) Referencing specific functions to implement functionality.
+The steps to implement are:
-(4) Change the format of data acquisition and save data or images.
+(1) Define the private function name and add the required parameters.
+(2) Use annotations to decorate the function.
+(3) Add the docstring to explain the use of this functionality.
+(4) Referencing specific libraries (e.g., Scikit-learn) to implement the functionality.
+(5) Change the format of data acquisition and save the produced data or images, etc.
+**2. Encapsulte the concrete code in Layer 1**
+Please refer to our framework's definition of **Layer 1** in section 1.
-***\*2. Define Common_component\****
+Some functions may use large code due to their complexity. To ensure the style and readability of the codebase, you need to put the specific function implementation into the corresponding `geochemistrypi/data_mining/model/func/mode/_common` files and call it.
-(1) Define the common_components in the parent class, its role is to set where the output is saved.
+The steps to implement are:
-(2) Set the parameter source for the added function.
+(1) Define the public function name, add the required parameters and proper decorator.
+(2) Add the docstring to explain the use of this functionality,the significance of each parameter and the related reference.
+(3) Implement functionality.
+(4) Returns the value used in **Layer 2**.
+**3. Define `common_components` Method**
+The steps to implement are:
-***\*3. Implement function functions\****
+(1) Define the path to store the data and images, etc.
+(2) Invoke the common application functions one by one.
-Some functions may use large code due to their complexity. To ensure the style and readability of the code, you need to put the specific function implementation into the corresponding `_common` files and call it.
+**4. Apeend The Name of Functionality in Class Attribute `common_function`**
-It includes:
+The steps to implement are:
-(1) Explain the significance of each parameter.
+(1) Create a class attribute `common_function` list in `ClusteringWorkflowBase`
+(2) Create a enum class to include the name of the functionality
+(3) Append the value of enum class into `common_function` list
-(2) Implement functionality.
+**Example**
-(3) Returns the required parameters.
+The following is the example of adding model evaluation score to the clustering base class.
+First, you need to find the base class of clustering.
+
-***\*eg:\**** You want to add model evaluation to your clustering.
+ -First, you need to find the parent class to clustering.
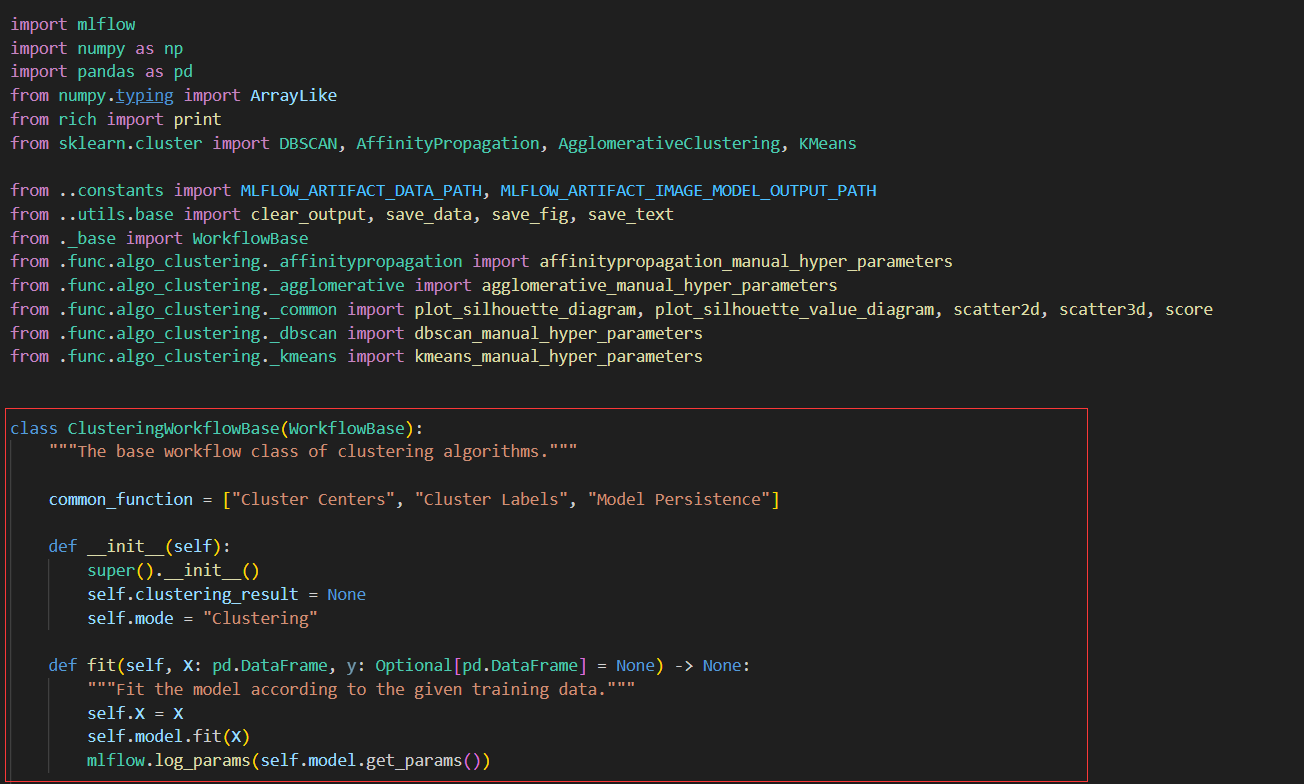
+**1. Add `_score` function in base class `ClusteringWorkflowBase(WorkflowBase)`**
-
+```python
+@staticmethod
+def _score(data: pd.DataFrame, labels: pd.DataFrame, func_name: str, algorithm_name: str, store_path: str) -> None:
+ """Calculate the score of the model."""
+ print(f"-----* {func_name} *-----")
+ scores = score(data, labels)
+ scores_str = json.dumps(scores, indent=4)
+ save_text(scores_str, f"{func_name}- {algorithm_name}", store_path)
+ mlflow.log_metrics(scores)
+```
-
+**2. Encapsulte the concrete code of `score` in Layer 1**
-***\*1. Add the clustering score function in class ClusteringWorkflowBase (WorkflowBase).\****
+You need to add the specific function implementation `score` to the corresponding `geochemistrypi/data_mining/model/func/algo_clustering/_common` file.
+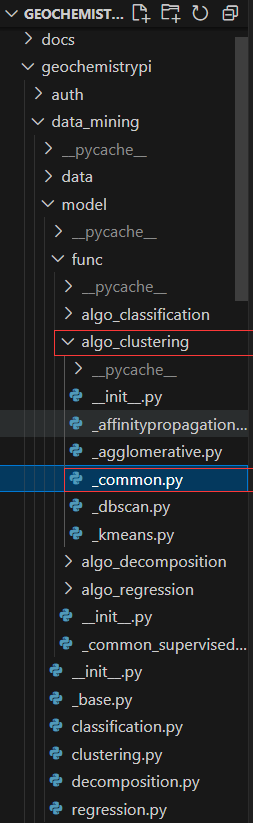
```python
+def score(data: pd.DataFrame, labels: pd.DataFrame) -> Dict:
+ """Calculate the scores of the clustering model.
-@staticmethod
-def _score(data: pd.DataFrame, labels: pd.DataFrame, algorithm_name: str, store_path: str) -> None:
+ Parameters
+ ----------
+ data : pd.DataFrame (n_samples, n_components)
+ The true values.
- """Calculate the score of the model."""
+ labels : pd.DataFrame (n_samples, n_components)
+ Labels of each point.
+
+ Returns
+ -------
+ scores : dict
+ The scores of the clustering model.
+ """
+ silhouette = silhouette_score(data, labels)
+ calinski_harabaz = calinski_harabasz_score(data, labels)
+ print("silhouette_score: ", silhouette)
+ print("calinski_harabasz_score:", calinski_harabaz)
+ scores = {
+ "silhouette_score": silhouette,
+ "calinski_harabasz_score": calinski_harabaz,
+ }
+ return scores
+```
- print("-----* Model Score *-----")
+**3. Define `common_components` Method in class `ClusteringWorkflowBase(WorkflowBase)`**
- scores = score(data, labels)
+```python
+def common_components(self) -> None:
+ """Invoke all common application functions for clustering algorithms."""
+ GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
+ GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
+ self._score(
+ data=self.X,
+ labels=self.clustering_result["clustering result"],
+ func_name=ClusteringCommonFunction.MODEL_SCORE.value,
+ algorithm_name=self.naming,
+ store_path=GEOPI_OUTPUT_METRICS_PATH,
+ )
+```
- scores_str = json.dumps(scores, indent=4)
+**4. Apeend The Name of Functionality in Class Attribute `common_function`**
- save_text(scores_str, f"Model Score - {algorithm_name}", store_path)
+Create a class attribute `common_function` in `ClusteringWorkflowBase`.
- mlflow.log_metrics(scores)
+```
+class ClusteringWorkflowBase(WorkflowBase):
+ """The base workflow class of clustering algorithms."""
+ common_function = [func.value for func in ClusteringCommonFunction]
```
+The enum class should be put in the corresponding path `geochemistrypi/data-mining/model/func/algo_clustering/_enum.py`
-(1) Define the function name and add the required parameters.
+
-First, you need to find the parent class to clustering.
+**1. Add `_score` function in base class `ClusteringWorkflowBase(WorkflowBase)`**
-
+```python
+@staticmethod
+def _score(data: pd.DataFrame, labels: pd.DataFrame, func_name: str, algorithm_name: str, store_path: str) -> None:
+ """Calculate the score of the model."""
+ print(f"-----* {func_name} *-----")
+ scores = score(data, labels)
+ scores_str = json.dumps(scores, indent=4)
+ save_text(scores_str, f"{func_name}- {algorithm_name}", store_path)
+ mlflow.log_metrics(scores)
+```
-
+**2. Encapsulte the concrete code of `score` in Layer 1**
-***\*1. Add the clustering score function in class ClusteringWorkflowBase (WorkflowBase).\****
+You need to add the specific function implementation `score` to the corresponding `geochemistrypi/data_mining/model/func/algo_clustering/_common` file.
+
```python
+def score(data: pd.DataFrame, labels: pd.DataFrame) -> Dict:
+ """Calculate the scores of the clustering model.
-@staticmethod
-def _score(data: pd.DataFrame, labels: pd.DataFrame, algorithm_name: str, store_path: str) -> None:
+ Parameters
+ ----------
+ data : pd.DataFrame (n_samples, n_components)
+ The true values.
- """Calculate the score of the model."""
+ labels : pd.DataFrame (n_samples, n_components)
+ Labels of each point.
+
+ Returns
+ -------
+ scores : dict
+ The scores of the clustering model.
+ """
+ silhouette = silhouette_score(data, labels)
+ calinski_harabaz = calinski_harabasz_score(data, labels)
+ print("silhouette_score: ", silhouette)
+ print("calinski_harabasz_score:", calinski_harabaz)
+ scores = {
+ "silhouette_score": silhouette,
+ "calinski_harabasz_score": calinski_harabaz,
+ }
+ return scores
+```
- print("-----* Model Score *-----")
+**3. Define `common_components` Method in class `ClusteringWorkflowBase(WorkflowBase)`**
- scores = score(data, labels)
+```python
+def common_components(self) -> None:
+ """Invoke all common application functions for clustering algorithms."""
+ GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
+ GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
+ self._score(
+ data=self.X,
+ labels=self.clustering_result["clustering result"],
+ func_name=ClusteringCommonFunction.MODEL_SCORE.value,
+ algorithm_name=self.naming,
+ store_path=GEOPI_OUTPUT_METRICS_PATH,
+ )
+```
- scores_str = json.dumps(scores, indent=4)
+**4. Apeend The Name of Functionality in Class Attribute `common_function`**
- save_text(scores_str, f"Model Score - {algorithm_name}", store_path)
+Create a class attribute `common_function` in `ClusteringWorkflowBase`.
- mlflow.log_metrics(scores)
+```
+class ClusteringWorkflowBase(WorkflowBase):
+ """The base workflow class of clustering algorithms."""
+ common_function = [func.value for func in ClusteringCommonFunction]
```
+The enum class should be put in the corresponding path `geochemistrypi/data-mining/model/func/algo_clustering/_enum.py`
-(1) Define the function name and add the required parameters.
+ -(2) Use annotations to describe function functionsUse annotations to describe function functions.
-(3) Referencing specific functions to implement functionality (Reference 3.2.3).
+#### 3.4.2 Add Special Application Functions and `special_components` Method
-(4) Change the format of data acquisition and save data or images.
+special application function is a feature that is unique to each specific model. The whole process is similar to that of previous sectoin for common functionalities.
-***\*Note:\**** Make sure that the code style of the added function is consistent.
+The process is as follows:
-***\*2. Define common_components below the added function to define the output position and parameter source for the added function.\****
+1. Add special application function with proper decorator to the child model workflow class
+2. Encapsulte the concrete code in Layer 1
+3. Define `special_components` method
+4. Apeend the name of functionality in class attribute `special_function`
-```python
+**Example**
-def common_components(self) -> None:
+Each algorithms has their own characteristics. Hence, they have different special fucntionalities as well. For example, for KMeans algorithm, we can get the inertia scores from it. Hence, inside the model workflow class `KMeansClustering`, we have a **private** instance method `_get_inertia_scores`.
- """Invoke all common application functions for clustering algorithms."""
+First, you need to find the child model workflow class for KMeans algorithm.
- GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
+
- GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
+
-(2) Use annotations to describe function functionsUse annotations to describe function functions.
-(3) Referencing specific functions to implement functionality (Reference 3.2.3).
+#### 3.4.2 Add Special Application Functions and `special_components` Method
-(4) Change the format of data acquisition and save data or images.
+special application function is a feature that is unique to each specific model. The whole process is similar to that of previous sectoin for common functionalities.
-***\*Note:\**** Make sure that the code style of the added function is consistent.
+The process is as follows:
-***\*2. Define common_components below the added function to define the output position and parameter source for the added function.\****
+1. Add special application function with proper decorator to the child model workflow class
+2. Encapsulte the concrete code in Layer 1
+3. Define `special_components` method
+4. Apeend the name of functionality in class attribute `special_function`
-```python
+**Example**
-def common_components(self) -> None:
+Each algorithms has their own characteristics. Hence, they have different special fucntionalities as well. For example, for KMeans algorithm, we can get the inertia scores from it. Hence, inside the model workflow class `KMeansClustering`, we have a **private** instance method `_get_inertia_scores`.
- """Invoke all common application functions for clustering algorithms."""
+First, you need to find the child model workflow class for KMeans algorithm.
- GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
+
- GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
+ - self._score(
+**1. Add `_get_inertia_scores` function in child model workflow class `KMeansClustering(ClusteringWorkflowBase)`**
- data=self.X,
+```python
+@staticmethod
+def _get_inertia_scores(func_name: str, algorithm_name: str, trained_model: object, store_path: str) -> None:
+ """Get the scores of the clustering result."""
+ print(f"-----* {func_name} *-----")
+ print(f"{func_name}: ", trained_model.inertia_)
+ inertia_scores = {f"{func_name}": trained_model.inertia_}
+ mlflow.log_metrics(inertia_scores)
+ inertia_scores_str = json.dumps(inertia_scores, indent=4)
+ save_text(inertia_scores_str, f"{func_name} - {algorithm_name}", store_path)
+```
- labels=self.clustering_result["clustering result"],
+**2. Encapsulte the concrete code in Layer 1**
- algorithm_name=self.naming,
+Getting the inertia score is only one line of code, hence no need to further encapsulate it.
- store_path=GEOPI_OUTPUT_METRICS_PATH,
+**3. Define `special_components` Method in class `KMeansClustering(ClusteringWorkflowBase)`**
+```python
+def special_components(self, **kwargs: Union[Dict, np.ndarray, int]) -> None:
+ """Invoke all special application functions for this algorithms by Scikit-learn framework."""
+ GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
+ self._get_inertia_scores(
+ func_name=KMeansSpecialFunction.INERTIA_SCORE.value,
+ algorithm_name=self.naming,
+ trained_model=self.model,
+ store_path=GEOPI_OUTPUT_METRICS_PATH,
)
-
```
-The positional relationship is shown in Figure 4.
+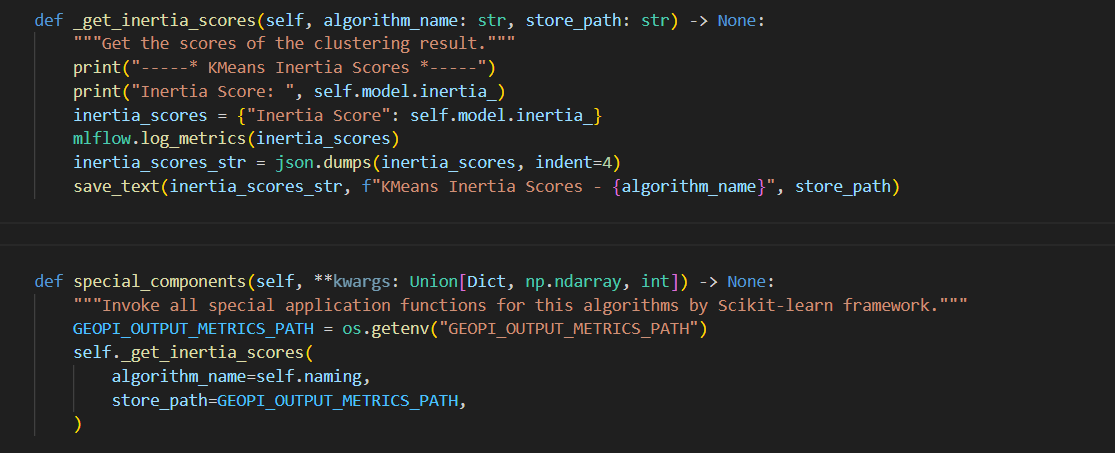
-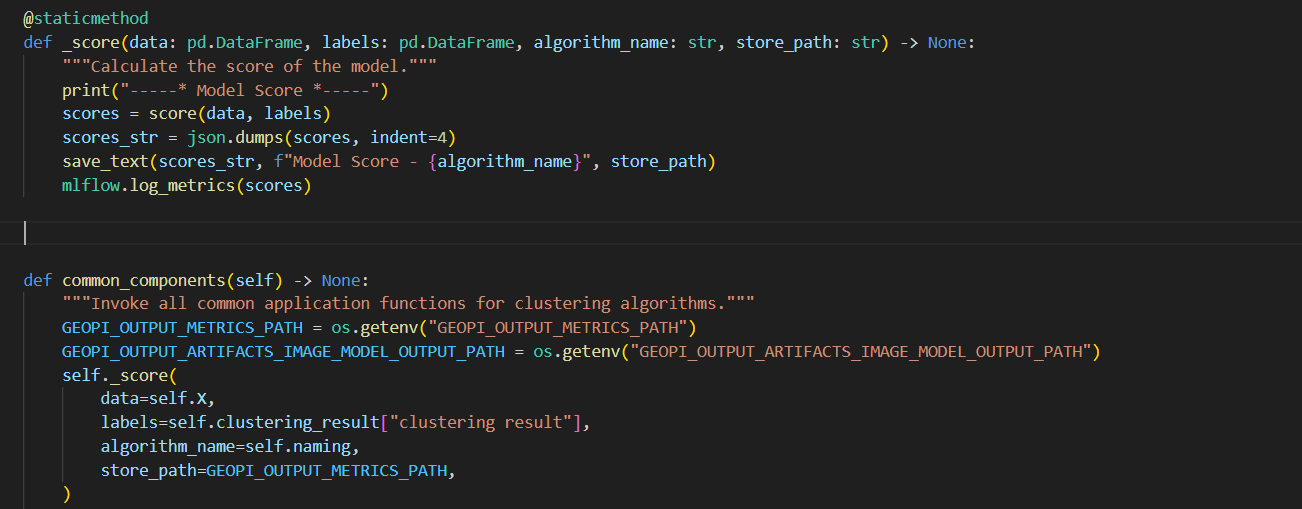
++ Also, if only part of the models share a functionality, for example, feature importance in tree-based algorithm including XGBoost, Decision Tree, etc. Hence, you can create a Mixin class to include that application function and let the tree-based model workflow class inherit it. Such as `ExtraTreesRegression(TreeWorkflowMixin, RegressionWorkflowBase)`
-***\*3. You need to add the specific function implementation to the corresponding `_commom` file.\****
+**4. Apeend The Name of Functionality in Class Attribute `special_function`**
-
+Create a class attribute `special_function` list in `KMeansClustering`.
-```python
+```
+class KMeansClustering(ClusteringWorkflowBase):
+ """The automation workflow of using KMeans algorithm to make insightful products."""
-def score(data: pd.DataFrame, labels: pd.DataFrame) -> Dict:
+ name = "KMeans"
+ special_function = [func.value for func in KMeansSpecialFunction]
+```
- """Calculate the scores of the clustering model.
+The enum class should be put in the corresponding path `geochemistrypi/data-mining/model/func/algo_clustering/_enum.py`
- Parameters
+
- self._score(
+**1. Add `_get_inertia_scores` function in child model workflow class `KMeansClustering(ClusteringWorkflowBase)`**
- data=self.X,
+```python
+@staticmethod
+def _get_inertia_scores(func_name: str, algorithm_name: str, trained_model: object, store_path: str) -> None:
+ """Get the scores of the clustering result."""
+ print(f"-----* {func_name} *-----")
+ print(f"{func_name}: ", trained_model.inertia_)
+ inertia_scores = {f"{func_name}": trained_model.inertia_}
+ mlflow.log_metrics(inertia_scores)
+ inertia_scores_str = json.dumps(inertia_scores, indent=4)
+ save_text(inertia_scores_str, f"{func_name} - {algorithm_name}", store_path)
+```
- labels=self.clustering_result["clustering result"],
+**2. Encapsulte the concrete code in Layer 1**
- algorithm_name=self.naming,
+Getting the inertia score is only one line of code, hence no need to further encapsulate it.
- store_path=GEOPI_OUTPUT_METRICS_PATH,
+**3. Define `special_components` Method in class `KMeansClustering(ClusteringWorkflowBase)`**
+```python
+def special_components(self, **kwargs: Union[Dict, np.ndarray, int]) -> None:
+ """Invoke all special application functions for this algorithms by Scikit-learn framework."""
+ GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
+ self._get_inertia_scores(
+ func_name=KMeansSpecialFunction.INERTIA_SCORE.value,
+ algorithm_name=self.naming,
+ trained_model=self.model,
+ store_path=GEOPI_OUTPUT_METRICS_PATH,
)
-
```
-The positional relationship is shown in Figure 4.
+
-
++ Also, if only part of the models share a functionality, for example, feature importance in tree-based algorithm including XGBoost, Decision Tree, etc. Hence, you can create a Mixin class to include that application function and let the tree-based model workflow class inherit it. Such as `ExtraTreesRegression(TreeWorkflowMixin, RegressionWorkflowBase)`
-***\*3. You need to add the specific function implementation to the corresponding `_commom` file.\****
+**4. Apeend The Name of Functionality in Class Attribute `special_function`**
-
+Create a class attribute `special_function` list in `KMeansClustering`.
-```python
+```
+class KMeansClustering(ClusteringWorkflowBase):
+ """The automation workflow of using KMeans algorithm to make insightful products."""
-def score(data: pd.DataFrame, labels: pd.DataFrame) -> Dict:
+ name = "KMeans"
+ special_function = [func.value for func in KMeansSpecialFunction]
+```
- """Calculate the scores of the clustering model.
+The enum class should be put in the corresponding path `geochemistrypi/data-mining/model/func/algo_clustering/_enum.py`
- Parameters
+ - ----------
- data : pd.DataFrame (n_samples, n_components)
+#### 3.4.3 Add `@dispatch()` to Component Method
- The true values.
+Howerever, in **regression** mode and **classification** mode, there are two different scenarios (AutoML and manual ML) when defining either `common_components` or `special_components` method. It is needed because we need to differentiate AutoML and manual ML. For example, inside the base model workflow class `RegressionWorkflowBase`, there are two `common_components` methods but with different decorators. Also, in its child model workflow class `ExtraTreesRegression`, there are two `special_components` methods but with different decorators.
- labels : pd.DataFrame (n_samples, n_components)
+Inside our framework, we leverages the thought of **method overloading** which is not supported by Python natively but we can achieve it through a library **multipledispatch**. The invocation of `common_components` and `special_components` method locates in Layer 3 which will be explained in later section.
- Labels of each point.
+The differences between AutoML and manual ML are as follows:
- Returns
+**1. The decorator**
- -------
++ For manual ML: add @dispatch() to decorate the component method
++ For AutoML: add @dispatch(bool) to decorate the component method
- scores : dict
+**2. The signature of the component method**
- The scores of the clustering model.
+For `common_compoents method`:
+```
+Manual ML:
+@dispatch()
+def common_components(self) -> None:
- """
+AutoML:
+@dispatch(bool)
+def common_components(self, is_automl: bool = False) -> None:
+```
- silhouette = silhouette_score(data, labels)
+For `special_compoents method`:
+```
+Manual ML:
+@dispatch()
+def special_components(self, **kwargs) -> None:
- calinski_harabaz = calinski_harabasz_score(data, labels)
+AutoML:
+@dispatch(bool)
+def special_components(self, is_automl: bool = False, **kwargs) -> None:
+```
- print("silhouette_score: ", silhouette)
+**3. The trained model instance variable**
- print("calinski_harabasz_score:", calinski_harabaz)
+Usually, inside the component method, we will pass the trained model instance variable to the application function. For example, for `common_components` in `RegressionWorkflowBase(WorkflowBase)`, be careful about the value passed to the parameter `trained_model`.
- scores = {
+```
+Manual ML:
+@dispatch()
+def common_components(self) -> None:
+ self._cross_validation(
+ trained_model=self.model,
+ X_train=RegressionWorkflowBase.X_train,
+ y_train=RegressionWorkflowBase.y_train,
+ cv_num=10,
+ algorithm_name=self.naming,
+ store_path=GEOPI_OUTPUT_METRICS_PATH,
+ )
+
+AutoML:
+@dispatch(bool)
+def common_components(self, is_automl: bool = False) -> None:
+ self._cross_validation(
+ trained_model=self.auto_model,
+ X_train=RegressionWorkflowBase.X_train,
+ y_train=RegressionWorkflowBase.y_train,
+ cv_num=10,
+ algorithm_name=self.naming,
+ store_path=GEOPI_OUTPUT_METRICS_PATH,
+)
+```
- "silhouette_score": silhouette,
+**Note:** The content of this part needs to be selected according to the actual situation of your own model. Can refer to similar classes.
- "calinski_harabasz_score": calinski_harabaz,
+#### 3.5 Storage Mechanism
- }
+In Geochemistry π, the storage mechanism consists of two components: the **geopi_tracking** folder and the **geopi_output** folder. MLflow uses the geopi_tracking folder as the store for visualized operation in the web interface, which researchers cannot modify directly. The geopi_output folder is a regular folder aligning with MLflow’s storage structure, which researchers can operate. Overall, this unique storage mechanism is purpose-built to track each experiment and its corresponding runs in order to create an organized and coherent record of researchers’ scientific explorations.
- return scores
+
- ----------
- data : pd.DataFrame (n_samples, n_components)
+#### 3.4.3 Add `@dispatch()` to Component Method
- The true values.
+Howerever, in **regression** mode and **classification** mode, there are two different scenarios (AutoML and manual ML) when defining either `common_components` or `special_components` method. It is needed because we need to differentiate AutoML and manual ML. For example, inside the base model workflow class `RegressionWorkflowBase`, there are two `common_components` methods but with different decorators. Also, in its child model workflow class `ExtraTreesRegression`, there are two `special_components` methods but with different decorators.
- labels : pd.DataFrame (n_samples, n_components)
+Inside our framework, we leverages the thought of **method overloading** which is not supported by Python natively but we can achieve it through a library **multipledispatch**. The invocation of `common_components` and `special_components` method locates in Layer 3 which will be explained in later section.
- Labels of each point.
+The differences between AutoML and manual ML are as follows:
- Returns
+**1. The decorator**
- -------
++ For manual ML: add @dispatch() to decorate the component method
++ For AutoML: add @dispatch(bool) to decorate the component method
- scores : dict
+**2. The signature of the component method**
- The scores of the clustering model.
+For `common_compoents method`:
+```
+Manual ML:
+@dispatch()
+def common_components(self) -> None:
- """
+AutoML:
+@dispatch(bool)
+def common_components(self, is_automl: bool = False) -> None:
+```
- silhouette = silhouette_score(data, labels)
+For `special_compoents method`:
+```
+Manual ML:
+@dispatch()
+def special_components(self, **kwargs) -> None:
- calinski_harabaz = calinski_harabasz_score(data, labels)
+AutoML:
+@dispatch(bool)
+def special_components(self, is_automl: bool = False, **kwargs) -> None:
+```
- print("silhouette_score: ", silhouette)
+**3. The trained model instance variable**
- print("calinski_harabasz_score:", calinski_harabaz)
+Usually, inside the component method, we will pass the trained model instance variable to the application function. For example, for `common_components` in `RegressionWorkflowBase(WorkflowBase)`, be careful about the value passed to the parameter `trained_model`.
- scores = {
+```
+Manual ML:
+@dispatch()
+def common_components(self) -> None:
+ self._cross_validation(
+ trained_model=self.model,
+ X_train=RegressionWorkflowBase.X_train,
+ y_train=RegressionWorkflowBase.y_train,
+ cv_num=10,
+ algorithm_name=self.naming,
+ store_path=GEOPI_OUTPUT_METRICS_PATH,
+ )
+
+AutoML:
+@dispatch(bool)
+def common_components(self, is_automl: bool = False) -> None:
+ self._cross_validation(
+ trained_model=self.auto_model,
+ X_train=RegressionWorkflowBase.X_train,
+ y_train=RegressionWorkflowBase.y_train,
+ cv_num=10,
+ algorithm_name=self.naming,
+ store_path=GEOPI_OUTPUT_METRICS_PATH,
+)
+```
- "silhouette_score": silhouette,
+**Note:** The content of this part needs to be selected according to the actual situation of your own model. Can refer to similar classes.
- "calinski_harabasz_score": calinski_harabaz,
+#### 3.5 Storage Mechanism
- }
+In Geochemistry π, the storage mechanism consists of two components: the **geopi_tracking** folder and the **geopi_output** folder. MLflow uses the geopi_tracking folder as the store for visualized operation in the web interface, which researchers cannot modify directly. The geopi_output folder is a regular folder aligning with MLflow’s storage structure, which researchers can operate. Overall, this unique storage mechanism is purpose-built to track each experiment and its corresponding runs in order to create an organized and coherent record of researchers’ scientific explorations.
- return scores
+
+ 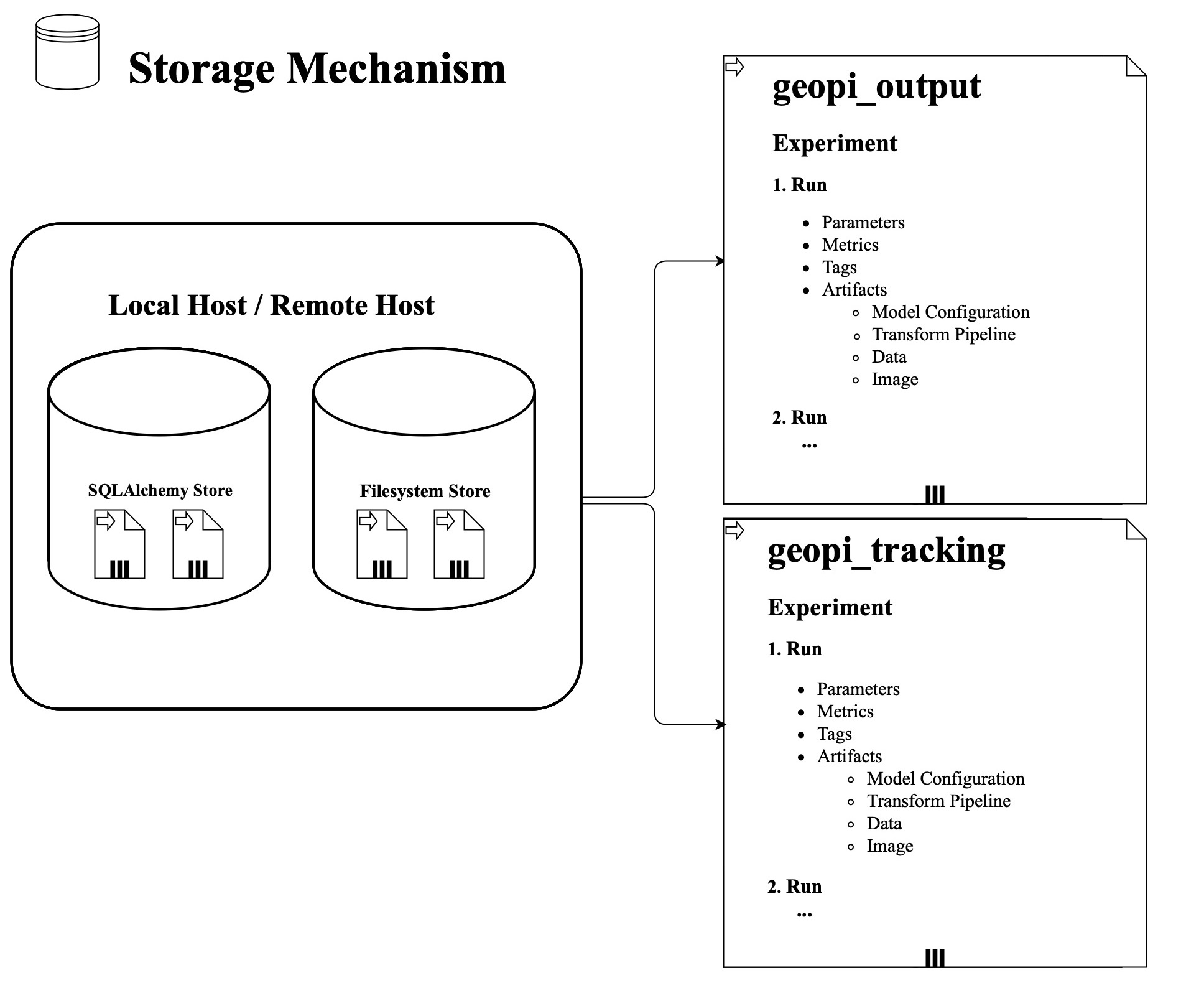 +
+
-```
+In the codebase, we use Python's open() function to store data into the **geopi_output** folder while MLflow's methods to store data into the **geopi_tracking** folder.
-(1) Explain the significance of each parameter.
+The common MLflow's methods includes:
-(2) Implement functionality.
++ mlflow.log_param(): Log a parameter (e.g. model hyperparameter) under the current run.
++ mlflow.log_params(): Log a batch of params for the current run.
++ mlflow.log_metric(): Log a metric under the current run.
++ mlflow.log_metrics(): Log multiple metrics for the current run.
++ mlflow.log_artifact(): Log a local file or directory as an artifact of the currently active run. In our software, we use it to store the images, data and text.
-(3) Returns the required parameters.
+You can refer the API document of MLflow for more details.
+Actually, we have encapsulated a bunch of saving functions in `geochemistrypi/data_mining/utils/base.py`, which can be used to store the data into the **geopi_output** folder and the **geopi_tracking** folder at the same time. It includes the functions `save_fig`, `save_data`, `save_text`, `save_model`.
+Usually, when you want to use the saving functions, you only need to pass it the storage path and data to store.
-#### 2.6.3 Add Special_component
+For example, in the case of adding a common application function into base clustering model workflow class.
-Special_components is a feature that is unique to each specific model.
+
-The process of adding a Special_components is similar to that of a Common_component.
+```
+GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
+```
++ This line of code gets the metrics output path from the environment variable.
+```
+GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
+```
++ This line of code gets the image model output path from the environment variable.
+```
+GEOPI_OUTPUT_ARTIFACTS_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_PATH")
+```
++ This line of code takes the general output artifact path from the environment variable.
+**Note:** You need to choose to add the corresponding path according to the usage in the following functions. You can look up the pre-defined pathes created inside the function `create_geopi_output_dir` in `geochemistrypi/data_mining/utils/base.py`.
+**Note:** You can refer to other similar model workflow classes to complete your implementation.
-The process is as follows:
-(1) Find the location that needs to be added.
+## 4. Instantiate Model Workflow Class
-(2) Defined function.
+### 4.1 Find File
-(3) Define Special_components and add a parametric function to it.
+Instantiating a model workflow class is the responsibilty of Layer 3. Layer 3 is represented by the scikit-learn API-style model selection interface in the corresponding mode file under the `geochemistrypi/data_mining/process` folder.
-(4) Add the corresponding specific function implementation function to the `corresponding manual parameter tuning` file.
+
+**eg:** If your model workflow class belongs to regression mode, you need to implement the creation of ML model workflow objects in `regress.py` file.
+### 4.2 Import Module
-***\*eg:\**** An example is to add a score evaluation function to k-means clustering.
+For example, for the model workflow class belonging to regression, you need to add your model inside `regress.py` file by using `from ..model.regression import()`.
-***\*1. Find the location that needs to be added.\****
+```
+from ..model.regression import(
+ ...
+ ModelWorkflowClass,
+)
+```
-We add his own unique score to the k-means.
+
-
+### 4.3 Define `activate` Method
-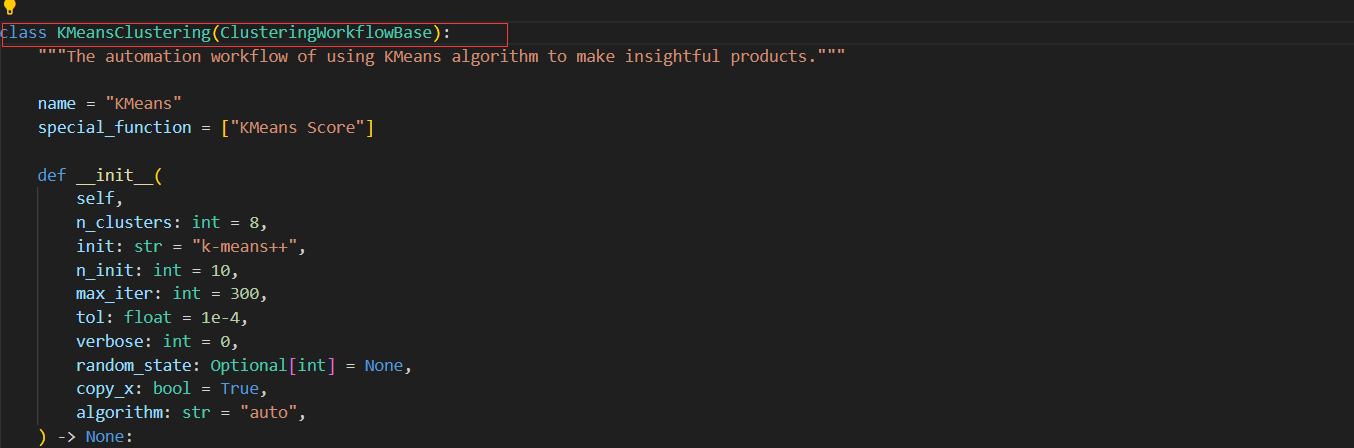
+The `activate` method defined in Layer 3 will be invoked in Layer 4.
-***\*2. Defined function.\****
+For supervised learning (regression and classification), the signature of `activate` method is:
+```
+def activate(
+ self,
+ X: pd.DataFrame,
+ y: pd.DataFrame,
+ X_train: pd.DataFrame,
+ X_test: pd.DataFrame,
+ y_train: pd.DataFrame,
+ y_test: pd.DataFrame,
+) -> None:
+ """Train by Scikit-learn framework."""
+```
-```python
+For unsupervised learning (clustering, decomposition and abnormaly detection), the signature of `activate` method is:
+```
+def activate(
+ self,
+ X: pd.DataFrame,
+ y: Optional[pd.DataFrame] = None,
+ X_train: Optional[pd.DataFrame] = None,
+ X_test: Optional[pd.DataFrame] = None,
+ y_train: Optional[pd.DataFrame] = None,
+ y_test: Optional[pd.DataFrame] = None,
+) -> None:
+ """Train by Scikit-learn framework."""
+```
-def _get_inertia_scores(self, algorithm_name: str, store_path: str) -> None:
+The difference is that for unsupervised learning, there is no need to seperate y and split the training-testing set. But for consistency, we keep it there.
- """Get the scores of the clustering result."""
+In **regression** mode and **classification** mode, there are two different scenarios (AutoML and manual ML) when defining either `activated` method. It is needed because we need to differentiate AutoML and manual ML. Hence, we still use @dispatch to decorate it. For example, in `RegressionModelSelection` class, we need to define two `activate` methods with different decorators.
- print("-----* KMeans Inertia Scores *-----")
+```
+Manual ML:
+@dispatch(object, object, object, object, object, object)
+def activate(
+ self,
+ X: pd.DataFrame,
+ y: pd.DataFrame,
+ X_train: pd.DataFrame,
+ X_test: pd.DataFrame,
+ y_train: pd.DataFrame,
+ y_test: pd.DataFrame,
+) -> None:
- print("Inertia Score: ", self.model.inertia_)
+AutoML:
+@dispatch(object, object, object, object, object, object, bool)
+def activate(
+ self,
+ X: pd.DataFrame,
+ y: pd.DataFrame,
+ X_train: pd.DataFrame,
+ X_test: pd.DataFrame,
+ y_train: pd.DataFrame,
+ y_test: pd.DataFrame,
+ is_automl: bool,
+) -> None:
+```
- inertia_scores = {"Inertia Score": self.model.inertia_}
+The differences above include the signature of @dispatch and the signature of `activate` method.
- mlflow.log_metrics(inertia_scores)
+### 4.4 Create Model Workflow Object
- inertia_scores_str = json.dumps(inertia_scores, indent=4)
+There are two `activate` methods defined in the Regression and Classification mode, the first method uses the Scikit-learn framework, and the second method uses the FLAML and RAY frameworks. Decomposition and Clustering algorithms only use the Scikit-learn framework. The instantiation of model workflow class inside `activate` method builds the connnectioni between Layer 3 and Layer 2.
- save_text(inertia_scores_str, f"KMeans Inertia Scores - {algorithm_name}", store_path)
+(1) The invocatioin of model workflow class in the first activate method (Used in classification, regression,decomposition, clustering, abnormaly detection) needs to pass the hyperparameters for manual ML:
+```
+elif self.model_name == "ModelName":
+ hyper_parameters = ModelWorkflowClass.manual_hyper_parameters()
+ self.dcp_workflow = ModelWorkflowClass(
+ Hyperparameters1=hyper_parameters["Hyperparameters2"],
+ Hyperparameters1=hyper_parameters["Hyperparameters2"],
+ ...
+ )
+```
++ This "ModelName" needs to be added to the corresponding constant variable in `geochemistrypi/data_mining/constants.py` file. It will be further explained in later section.
+
+**eg:**
+
+
+(2)The invocatioin of model workflow class in the second activate method(Used in classification, regression)for AutoML:
```
+elif self.model_name == "ModelName":
+ self.reg_workflow = ModelWorkflowClass()
+```
++ This "ModelName" needs to be added to the corresponding constant variable in `geochemistrypi/data_mining/constants.py` file. It will be further explained in later section.
-(1) Define the function name and add the required parameters.
+**eg:**
+
-(2) Use annotations to describe function functionsUse annotations to describe function functions.
+### 4.5 Invoke Other Methods in Scikit-learn API Style
-(3) Referencing specific functions to implement functionality.
+It should contain at least these functoins below:
-(4) Change the format of data acquisition and save data or images.
++ data_upload(): Load the required data into the base class's attributes.
++ show_info(): Display what application functions the algorithm will provide.
++ fit(): Fit the model.
++ save_hyper_parameters(): Save the model hyper-parameters into the storage.
++ common_components(): Invoke all common application functions.
++ special_components(): Invoke all special application functions.
++ model_save(): Save the trained model.
-***\*3. Define Special_components and add a parametric function to it.\****
+You can refer to other existing mode inside `geochemistrypi/data_mining/process/mode.py` to see what other else you need.
+### 4.6 Add `model_name` to `MODE_MODELS` or `NON_AUTOML_MODELS`
-```python
+Find the `constants.py` file under `geochemistrypi/data_mining` folder to add the model name which should be identical to that in `geochemistrypi/data_mining/process/mode.py` and in `geochemistrypi/data_mining/model/mode.py`.
-def special_components(self, **kwargs: Union[Dict, np.ndarray, int]) -> None:
+
- """Invoke all special application functions for this algorithms by Scikit-learn framework."""
+**(1) Add `model_name` to `MODE_MODELS`**
- GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
+Append `model_name` to the `MODE_MODELS` list corresponding to the mode in the constants file.
- self._get_inertia_scores(
+**eg:** Add the name of the Lasso regression algorithm to `REGRESSION_MODELS` list.
- algorithm_name=self.naming,
+
- store_path=GEOPI_OUTPUT_METRICS_PATH,
+**(2) Add `model_name` to `NON_AUTOML_MODELS`**
- )
+Only for those algorithms, they belong to either regression or classification and don't need to provide AutoML functionality. They need to append `model_name` to `NON_AUTOML_MODELS` list.
-```
+**eg:** Add the name of the Linear Regression algorithm to `NON_AUTOML_MODELS` list.
-The positional relationship is shown in Figure 7.
+
-
-***\*4. Add the corresponding specific function implementation function to the `corresponding manual parameter tuning` file.\****
+## 5. Test Model Workflow Class
+
+After the model workflow class is added, you can test it through running the command `python start_cli_pipeline.py` on the terminal. If the test reports an error, you need to debug and fix it. If there is no error, it can be submitted.
-If the defined function has complex functions, it is necessary to further improve its function content in the manual parameter file, and the code format should refer to Common_component.
-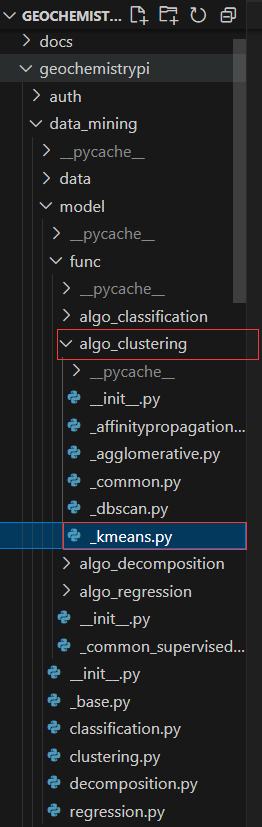
+## 6. Completed Pull Request
-## 3. Test model
-After the model is added, it can be tested. If the test reports an error, it needs to be checked. If there is no error, it can be submitted.
+After the test is correct, you can complete the pull request according to the puu document instructions in [Geochemistry π - Completed Pull Request](https://geochemistrypi.readthedocs.io/en/latest/index.html)
-## 4. Completed Pull Request
-After the model test is correct, you can complete the pull request according to the puu document instructions in [Geochemistry π](https://geochemistrypi.readthedocs.io/en/latest/index.html)
-## 5. Precautions
+
+## 7. Precautions
**Note1:** This tutorial only discusses the general process of adding a model, and the specific addition needs to be combined with the actual situation of the model to accurately add relevant codes.
**Note2:** If there are unclear situations and problems during the adding process, communicate with other people in time to solve them.
diff --git a/geochemistrypi/data_mining/model/_base.py b/geochemistrypi/data_mining/model/_base.py
index c57d14c8..8e701a3f 100644
--- a/geochemistrypi/data_mining/model/_base.py
+++ b/geochemistrypi/data_mining/model/_base.py
@@ -30,7 +30,7 @@ class WorkflowBase(metaclass=ABCMeta):
@classmethod
def show_info(cls) -> None:
- """Display how many functions the algorithm will provide."""
+ """Display what application functions the algorithm will provide."""
print("*-*" * 2, cls.name, "is running ...", "*-*" * 2)
print("Expected Functionality:")
function = cls.common_function + cls.special_function
diff --git a/geochemistrypi/data_mining/model/clustering.py b/geochemistrypi/data_mining/model/clustering.py
index 6dc456ae..5a8b34d0 100644
--- a/geochemistrypi/data_mining/model/clustering.py
+++ b/geochemistrypi/data_mining/model/clustering.py
@@ -17,13 +17,14 @@
from .func.algo_clustering._agglomerative import agglomerative_manual_hyper_parameters
from .func.algo_clustering._common import plot_silhouette_diagram, plot_silhouette_value_diagram, scatter2d, scatter3d, score
from .func.algo_clustering._dbscan import dbscan_manual_hyper_parameters
+from .func.algo_clustering._enum import ClusteringCommonFunction, KMeansSpecialFunction
from .func.algo_clustering._kmeans import kmeans_manual_hyper_parameters
class ClusteringWorkflowBase(WorkflowBase):
"""The base workflow class of clustering algorithms."""
- common_function = ["Cluster Centers", "Cluster Labels", "Model Persistence"]
+ common_function = [func.value for func in ClusteringCommonFunction]
def __init__(self):
super().__init__()
@@ -58,12 +59,12 @@ def get_labels(self):
save_data(self.clustering_result, f"{self.naming} Result", GEOPI_OUTPUT_ARTIFACTS_DATA_PATH, MLFLOW_ARTIFACT_DATA_PATH)
@staticmethod
- def _score(data: pd.DataFrame, labels: pd.DataFrame, algorithm_name: str, store_path: str) -> None:
+ def _score(data: pd.DataFrame, labels: pd.DataFrame, func_name: str, algorithm_name: str, store_path: str) -> None:
"""Calculate the score of the model."""
- print("-----* Model Score *-----")
+ print(f"-----* {func_name} *-----")
scores = score(data, labels)
scores_str = json.dumps(scores, indent=4)
- save_text(scores_str, f"Model Score - {algorithm_name}", store_path)
+ save_text(scores_str, f"{func_name}- {algorithm_name}", store_path)
mlflow.log_metrics(scores)
@staticmethod
@@ -112,6 +113,7 @@ def common_components(self) -> None:
self._score(
data=self.X,
labels=self.clustering_result["clustering result"],
+ func_name=ClusteringCommonFunction.MODEL_SCORE.value,
algorithm_name=self.naming,
store_path=GEOPI_OUTPUT_METRICS_PATH,
)
@@ -190,7 +192,7 @@ class KMeansClustering(ClusteringWorkflowBase):
"""The automation workflow of using KMeans algorithm to make insightful products."""
name = "KMeans"
- special_function = ["KMeans Score"]
+ special_function = [func.value for func in KMeansSpecialFunction]
def __init__(
self,
@@ -304,14 +306,15 @@ def __init__(
self.naming = KMeansClustering.name
- def _get_inertia_scores(self, algorithm_name: str, store_path: str) -> None:
+ @staticmethod
+ def _get_inertia_scores(func_name: str, algorithm_name: str, trained_model: object, store_path: str) -> None:
"""Get the scores of the clustering result."""
- print("-----* KMeans Inertia Scores *-----")
- print("Inertia Score: ", self.model.inertia_)
- inertia_scores = {"Inertia Score": self.model.inertia_}
+ print(f"-----* {func_name} *-----")
+ print(f"{func_name}: ", trained_model.inertia_)
+ inertia_scores = {f"{func_name}": trained_model.inertia_}
mlflow.log_metrics(inertia_scores)
inertia_scores_str = json.dumps(inertia_scores, indent=4)
- save_text(inertia_scores_str, f"KMeans Inertia Scores - {algorithm_name}", store_path)
+ save_text(inertia_scores_str, f"{func_name} - {algorithm_name}", store_path)
@classmethod
def manual_hyper_parameters(cls) -> Dict:
@@ -325,7 +328,9 @@ def special_components(self, **kwargs: Union[Dict, np.ndarray, int]) -> None:
"""Invoke all special application functions for this algorithms by Scikit-learn framework."""
GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
self._get_inertia_scores(
+ func_name=KMeansSpecialFunction.INERTIA_SCORE.value,
algorithm_name=self.naming,
+ trained_model=self.model,
store_path=GEOPI_OUTPUT_METRICS_PATH,
)
diff --git a/geochemistrypi/data_mining/model/func/algo_clustering/_enum.py b/geochemistrypi/data_mining/model/func/algo_clustering/_enum.py
new file mode 100644
index 00000000..22293cac
--- /dev/null
+++ b/geochemistrypi/data_mining/model/func/algo_clustering/_enum.py
@@ -0,0 +1,12 @@
+from enum import Enum
+
+
+class ClusteringCommonFunction(Enum):
+ CLUSTER_CENTERS = "Cluster Centers"
+ CLUSTER_LABELS = "Cluster Labels"
+ MODEL_PERSISTENCE = "Model Persistence"
+ MODEL_SCORE = "Model Score"
+
+
+class KMeansSpecialFunction(Enum):
+ INERTIA_SCORE = "Inertia Score"
diff --git a/geochemistrypi/data_mining/model/regression.py b/geochemistrypi/data_mining/model/regression.py
index e78cda95..06a33c5b 100644
--- a/geochemistrypi/data_mining/model/regression.py
+++ b/geochemistrypi/data_mining/model/regression.py
@@ -200,7 +200,7 @@ def common_components(self) -> None:
)
@dispatch(bool)
- def common_components(self, is_automl: bool) -> None:
+ def common_components(self, is_automl: bool = False) -> None:
"""Invoke all common application functions for regression algorithms by FLAML framework."""
GEOPI_OUTPUT_METRICS_PATH = os.getenv("GEOPI_OUTPUT_METRICS_PATH")
GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
@@ -904,7 +904,7 @@ def manual_hyper_parameters(cls) -> Dict:
return hyper_parameters
@dispatch()
- def special_components(self):
+ def special_components(self, **kwargs):
"""Invoke all special application functions for this algorithms by Scikit-learn framework."""
GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH = os.getenv("GEOPI_OUTPUT_ARTIFACTS_IMAGE_MODEL_OUTPUT_PATH")
self._plot_feature_importance(
diff --git a/geochemistrypi/data_mining/process/classify.py b/geochemistrypi/data_mining/process/classify.py
index 42768353..324cb591 100644
--- a/geochemistrypi/data_mining/process/classify.py
+++ b/geochemistrypi/data_mining/process/classify.py
@@ -41,6 +41,7 @@ def activate(
) -> None:
"""Train by Scikit-learn framework."""
+ # Load the required data into the base class's attributes
self.clf_workflow.data_upload(X=X, y=y, X_train=X_train, X_test=X_test, y_train=y_train, y_test=y_test)
# Customize label
@@ -163,6 +164,7 @@ def activate(
validation_fraction=hyper_parameters["validation_fraction"],
n_iter_no_change=hyper_parameters["n_iter_no_change"],
)
+ # Display what application functions the algorithm will provide
self.clf_workflow.show_info()
# Use Scikit-learn style API to process input data
diff --git a/geochemistrypi/data_mining/process/detect.py b/geochemistrypi/data_mining/process/detect.py
index cf49df5d..94313758 100644
--- a/geochemistrypi/data_mining/process/detect.py
+++ b/geochemistrypi/data_mining/process/detect.py
@@ -16,7 +16,6 @@ def __init__(self, model_name: str) -> None:
self.ad_workflow = AbnormalDetectionWorkflowBase()
self.transformer_config = {}
- # @dispatch(object, object, object, object, object, object)
def activate(
self,
X: pd.DataFrame,
+
+